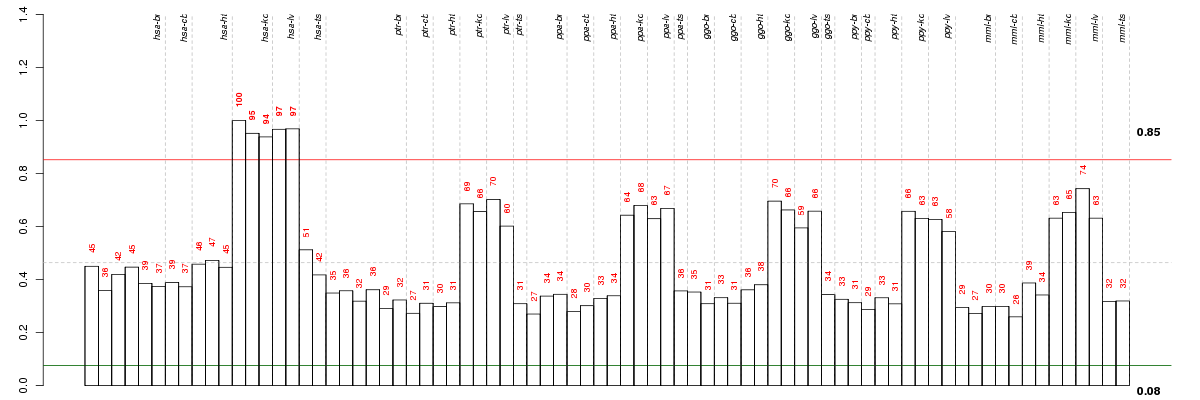

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
pyrimidine base metabolic process
The chemical reactions and pathways involving pyrimidine bases, 1,3-diazine, organic nitrogenous bases.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
cellular aromatic compound metabolic process
The chemical reactions and pathways involving aromatic compounds, any organic compound characterized by one or more planar rings, each of which contains conjugated double bonds and delocalized pi electrons, as carried out by individual cells.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
homophilic cell adhesion
The attachment of an adhesion molecule in one cell to an identical molecule in an adjacent cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
nucleobase metabolic process
The chemical reactions and pathways involving a nucleobase, a nitrogenous base that is a constituent of a nucleic acid, e.g. the purines: adenine, guanine, hypoxanthine, xanthine and the pyrimidines: cytosine, uracil, thymine.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
heterocycle metabolic process
The chemical reactions and pathways involving heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
nucleobase metabolic process
The chemical reactions and pathways involving a nucleobase, a nitrogenous base that is a constituent of a nucleic acid, e.g. the purines: adenine, guanine, hypoxanthine, xanthine and the pyrimidines: cytosine, uracil, thymine.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
nucleobase metabolic process
The chemical reactions and pathways involving a nucleobase, a nitrogenous base that is a constituent of a nucleic acid, e.g. the purines: adenine, guanine, hypoxanthine, xanthine and the pyrimidines: cytosine, uracil, thymine.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.

calcium ion binding
Interacting selectively and non-covalently with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively and non-covalently with any metal ion.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which an aldehyde or ketone (oxo) group acts as a hydrogen or electron donor and reduces NAD or NADP.
oxidoreductase activity, acting on the aldehyde or oxo group of donors
Catalysis of an oxidation-reduction (redox) reaction in which an aldehyde or ketone (oxo) group acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
lyase activity
Catalysis of the cleavage of C-C, C-O, C-N and other bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond. They differ from other enzymes in that two substrates are involved in one reaction direction, but only one in the other direction. When acting on the single substrate, a molecule is eliminated and this generates either a new double bond or a new ring.
carbon-carbon lyase activity
Catalysis of the cleavage of C-C bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond.
ion binding
Interacting selectively and non-covalently with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively and non-covalently with cations, charged atoms or groups of atoms with a net positive charge.
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
coenzyme binding
Interacting selectively and non-covalently with a coenzyme, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 01100 | 2.294e-05 | 33.79 | 61 | 805 | Metabolic pathways |
| 04146 | 2.644e-03 | 2.812 | 11 | 67 | Peroxisome |
| 00340 | 8.696e-03 | 1.007 | 6 | 24 | Histidine metabolism |
| 00010 | 2.937e-02 | 1.763 | 7 | 42 | Glycolysis / Gluconeogenesis |
| 00040 | 3.899e-02 | 0.2938 | 3 | 7 | Pentose and glucuronate interconversions |
| 00360 | 4.739e-02 | 0.6296 | 4 | 15 | Phenylalanine metabolism |
AAMPangio-associated, migratory cell protein (ENSG00000127837), score: 0.5 ABHD14Babhydrolase domain containing 14B (ENSG00000114779), score: 0.47 ACAA1acetyl-CoA acyltransferase 1 (ENSG00000060971), score: 0.48 ACAD10acyl-CoA dehydrogenase family, member 10 (ENSG00000111271), score: 0.55 ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.5 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.52 ACOT11acyl-CoA thioesterase 11 (ENSG00000162390), score: 0.47 ACSS2acyl-CoA synthetase short-chain family member 2 (ENSG00000131069), score: 0.5 ACY3aspartoacylase (aminocyclase) 3 (ENSG00000132744), score: 0.51 AFMIDarylformamidase (ENSG00000183077), score: 0.5 AGMATagmatine ureohydrolase (agmatinase) (ENSG00000116771), score: 0.49 AGPAT21-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) (ENSG00000169692), score: 0.48 AGPAT31-acylglycerol-3-phosphate O-acyltransferase 3 (ENSG00000160216), score: 0.49 AGXT2alanine--glyoxylate aminotransferase 2 (ENSG00000113492), score: 0.49 AGXT2L2alanine-glyoxylate aminotransferase 2-like 2 (ENSG00000175309), score: 0.54 AHNAK2AHNAK nucleoprotein 2 (ENSG00000185567), score: -0.49 AIG1androgen-induced 1 (ENSG00000146416), score: 0.46 AIMP2aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (ENSG00000106305), score: 0.53 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.49 ALDH16A1aldehyde dehydrogenase 16 family, member A1 (ENSG00000161618), score: 0.48 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (ENSG00000137124), score: 0.46 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (ENSG00000006534), score: 0.47 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (ENSG00000159423), score: 0.53 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (ENSG00000119711), score: 0.46 ALDH8A1aldehyde dehydrogenase 8 family, member A1 (ENSG00000118514), score: 0.48 ALDOBaldolase B, fructose-bisphosphate (ENSG00000136872), score: 0.52 ANKS4Bankyrin repeat and sterile alpha motif domain containing 4B (ENSG00000175311), score: 0.49 ANPEPalanyl (membrane) aminopeptidase (ENSG00000166825), score: 0.49 ANXA13annexin A13 (ENSG00000104537), score: 0.54 ANXA9annexin A9 (ENSG00000143412), score: 0.57 AP4B1adaptor-related protein complex 4, beta 1 subunit (ENSG00000134262), score: -0.5 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.59 APEX1APEX nuclease (multifunctional DNA repair enzyme) 1 (ENSG00000100823), score: 0.57 APEX2APEX nuclease (apurinic/apyrimidinic endonuclease) 2 (ENSG00000169188), score: 0.52 APOMapolipoprotein M (ENSG00000204444), score: 0.47 AQP3aquaporin 3 (Gill blood group) (ENSG00000165272), score: 0.64 ARHGEF2Rho/Rac guanine nucleotide exchange factor (GEF) 2 (ENSG00000116584), score: -0.49 ARSAarylsulfatase A (ENSG00000100299), score: 0.47 ARSEarylsulfatase E (chondrodysplasia punctata 1) (ENSG00000157399), score: 0.72 AS3MTarsenic (+3 oxidation state) methyltransferase (ENSG00000214435), score: 0.49 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.48 ASPDHaspartate dehydrogenase domain containing (ENSG00000204653), score: 0.49 ASPGasparaginase homolog (S. cerevisiae) (ENSG00000166183), score: 0.58 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.6 ATP12AATPase, H+/K+ transporting, nongastric, alpha polypeptide (ENSG00000075673), score: 0.54 B3GNT3UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 (ENSG00000179913), score: 0.8 BAT5HLA-B associated transcript 5 (ENSG00000204422), score: 0.53 BBOX1butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 (ENSG00000129151), score: 0.48 BBS7Bardet-Biedl syndrome 7 (ENSG00000138686), score: -0.52 BCKDKbranched chain ketoacid dehydrogenase kinase (ENSG00000103507), score: 0.53 BHMT2betaine--homocysteine S-methyltransferase 2 (ENSG00000132840), score: 0.52 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.5 BOD1Lbiorientation of chromosomes in cell division 1-like (ENSG00000038219), score: -0.55 BPHLbiphenyl hydrolase-like (serine hydrolase) (ENSG00000137274), score: 0.46 BPTFbromodomain PHD finger transcription factor (ENSG00000171634), score: -0.49 C11orf52chromosome 11 open reading frame 52 (ENSG00000149300), score: 0.47 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.54 C12orf4chromosome 12 open reading frame 4 (ENSG00000047621), score: -0.52 C12orf52chromosome 12 open reading frame 52 (ENSG00000139405), score: 0.52 C14orf21chromosome 14 open reading frame 21 (ENSG00000196943), score: 0.55 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: 0.65 C16orf58chromosome 16 open reading frame 58 (ENSG00000140688), score: 0.55 C17orf106chromosome 17 open reading frame 106 (ENSG00000108504), score: 0.51 C17orf106chromosome 17 open reading frame 106 (ENSG00000214143), score: 0.49 C19orf39chromosome 19 open reading frame 39 (ENSG00000173928), score: 0.51 C1orf186chromosome 1 open reading frame 186 (ENSG00000196533), score: 0.92 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: 0.59 C1orf52chromosome 1 open reading frame 52 (ENSG00000162642), score: -0.51 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.49 C22orf13chromosome 22 open reading frame 13 (ENSG00000138867), score: 0.48 C22orf28chromosome 22 open reading frame 28 (ENSG00000100220), score: 0.53 C2orf24chromosome 2 open reading frame 24 (ENSG00000115649), score: 0.47 C2orf49chromosome 2 open reading frame 49 (ENSG00000135974), score: -0.55 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.54 C2orf79chromosome 2 open reading frame 79 (ENSG00000184924), score: 0.49 C5orf15chromosome 5 open reading frame 15 (ENSG00000113583), score: 0.59 C5orf45chromosome 5 open reading frame 45 (ENSG00000161010), score: 0.49 C6orf201chromosome 6 open reading frame 201 (ENSG00000185689), score: 0.46 C8orf55chromosome 8 open reading frame 55 (ENSG00000130193), score: 0.48 C9orf129chromosome 9 open reading frame 129 (ENSG00000204352), score: -0.52 CAPN12calpain 12 (ENSG00000182472), score: 0.47 CBLCCas-Br-M (murine) ecotropic retroviral transforming sequence c (ENSG00000142273), score: 0.58 CBR4carbonyl reductase 4 (ENSG00000145439), score: 0.49 CCDC90Acoiled-coil domain containing 90A (ENSG00000050393), score: 0.46 CCNB1IP1cyclin B1 interacting protein 1 (ENSG00000100814), score: 0.54 CCNT2cyclin T2 (ENSG00000082258), score: -0.59 CDAcytidine deaminase (ENSG00000158825), score: 0.47 CDC7cell division cycle 7 homolog (S. cerevisiae) (ENSG00000097046), score: -0.48 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.48 CDHR2cadherin-related family member 2 (ENSG00000074276), score: 0.71 CDHR5cadherin-related family member 5 (ENSG00000099834), score: 0.67 CEACAM1carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) (ENSG00000079385), score: 0.48 CECR5cat eye syndrome chromosome region, candidate 5 (ENSG00000069998), score: 0.58 CGREF1cell growth regulator with EF-hand domain 1 (ENSG00000138028), score: 0.48 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: 0.57 CHD6chromodomain helicase DNA binding protein 6 (ENSG00000124177), score: -0.5 CHEK2CHK2 checkpoint homolog (S. pombe) (ENSG00000183765), score: 0.49 CHID1chitinase domain containing 1 (ENSG00000177830), score: 0.53 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (ENSG00000110172), score: -0.48 CHP2calcineurin B homologous protein 2 (ENSG00000166869), score: 0.47 CHST13carbohydrate (chondroitin 4) sulfotransferase 13 (ENSG00000180767), score: 0.47 CIB2calcium and integrin binding family member 2 (ENSG00000136425), score: -0.53 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.52 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.56 CITED4Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 (ENSG00000179862), score: 0.52 CLCN5chloride channel 5 (ENSG00000171365), score: 0.52 CLDN3claudin 3 (ENSG00000165215), score: 0.49 CLN5ceroid-lipofuscinosis, neuronal 5 (ENSG00000102805), score: 0.47 CLN6ceroid-lipofuscinosis, neuronal 6, late infantile, variant (ENSG00000128973), score: 0.59 CLPTM1LCLPTM1-like (ENSG00000049656), score: 0.58 CLRN3clarin 3 (ENSG00000180745), score: 0.69 CLYBLcitrate lyase beta like (ENSG00000125246), score: 0.48 CNDP2CNDP dipeptidase 2 (metallopeptidase M20 family) (ENSG00000133313), score: 0.5 CNGA1cyclic nucleotide gated channel alpha 1 (ENSG00000198515), score: 0.58 COASYCoA synthase (ENSG00000068120), score: 0.58 COPZ1coatomer protein complex, subunit zeta 1 (ENSG00000111481), score: 0.51 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.61 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (ENSG00000014919), score: 0.55 CPEB4cytoplasmic polyadenylation element binding protein 4 (ENSG00000113742), score: -0.51 CPN2carboxypeptidase N, polypeptide 2 (ENSG00000178772), score: 0.53 CRB3crumbs homolog 3 (Drosophila) (ENSG00000130545), score: 0.51 CRYAAcrystallin, alpha A (ENSG00000160202), score: 0.65 CRYBB3crystallin, beta B3 (ENSG00000100053), score: 0.64 CRYL1crystallin, lambda 1 (ENSG00000165475), score: 0.48 CSTAcystatin A (stefin A) (ENSG00000121552), score: 0.48 CTAGE5CTAGE family, member 5 (ENSG00000150527), score: 0.49 CTRCchymotrypsin C (caldecrin) (ENSG00000162438), score: 0.47 CTSDcathepsin D (ENSG00000117984), score: 0.49 CTSHcathepsin H (ENSG00000103811), score: 0.47 CUTAcutA divalent cation tolerance homolog (E. coli) (ENSG00000112514), score: 0.53 CWH43cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) (ENSG00000109182), score: 0.56 CYB561D2cytochrome b-561 domain containing 2 (ENSG00000114395), score: 0.5 CYB5D2cytochrome b5 domain containing 2 (ENSG00000167740), score: 0.47 CYBASC3cytochrome b, ascorbate dependent 3 (ENSG00000162144), score: 0.48 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.5 CYP27B1cytochrome P450, family 27, subfamily B, polypeptide 1 (ENSG00000111012), score: 0.5 DACT2dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis) (ENSG00000164488), score: 0.46 DAKdihydroxyacetone kinase 2 homolog (S. cerevisiae) (ENSG00000149476), score: 0.51 DAOD-amino-acid oxidase (ENSG00000110887), score: 0.48 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.46 DCTN4dynactin 4 (p62) (ENSG00000132912), score: -0.49 DCXRdicarbonyl/L-xylulose reductase (ENSG00000169738), score: 0.53 DDCdopa decarboxylase (aromatic L-amino acid decarboxylase) (ENSG00000132437), score: 0.48 DDHD1DDHD domain containing 1 (ENSG00000100523), score: -0.48 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (ENSG00000100201), score: -0.49 DDX26BDEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B (ENSG00000165359), score: -0.5 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (ENSG00000123064), score: 0.47 DEFB1defensin, beta 1 (ENSG00000164825), score: 0.49 DENND1CDENN/MADD domain containing 1C (ENSG00000205744), score: 0.52 DENND2DDENN/MADD domain containing 2D (ENSG00000162777), score: 0.48 DEPDC6DEP domain containing 6 (ENSG00000155792), score: 0.48 DERAdeoxyribose-phosphate aldolase (putative) (ENSG00000023697), score: 0.51 DHRS2dehydrogenase/reductase (SDR family) member 2 (ENSG00000100867), score: 0.62 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.55 DIAPH1diaphanous homolog 1 (Drosophila) (ENSG00000131504), score: 0.47 DIO1deiodinase, iodothyronine, type I (ENSG00000211452), score: 0.48 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (ENSG00000160305), score: -0.57 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.72 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.49 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.52 DPYSdihydropyrimidinase (ENSG00000147647), score: 0.52 DUSP23dual specificity phosphatase 23 (ENSG00000158716), score: 0.57 ECHDC2enoyl CoA hydratase domain containing 2 (ENSG00000121310), score: 0.47 EEF1Geukaryotic translation elongation factor 1 gamma (ENSG00000149016), score: 0.67 EEF2eukaryotic translation elongation factor 2 (ENSG00000167658), score: 0.47 EIF3Ieukaryotic translation initiation factor 3, subunit I (ENSG00000084623), score: 0.49 EIF3Keukaryotic translation initiation factor 3, subunit K (ENSG00000178982), score: 0.53 EIF4E2eukaryotic translation initiation factor 4E family member 2 (ENSG00000135930), score: 0.47 ELAC2elaC homolog 2 (E. coli) (ENSG00000006744), score: 0.52 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.75 EPC2enhancer of polycomb homolog 2 (Drosophila) (ENSG00000135999), score: -0.51 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.5 ERGIC1endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 (ENSG00000113719), score: 0.6 ERP27endoplasmic reticulum protein 27 (ENSG00000139055), score: 0.54 ERP29endoplasmic reticulum protein 29 (ENSG00000089248), score: 0.49 ETNK2ethanolamine kinase 2 (ENSG00000143845), score: 0.46 EXOC1exocyst complex component 1 (ENSG00000090989), score: -0.51 EXOGendo/exonuclease (5'-3'), endonuclease G-like (ENSG00000157036), score: -0.49 FAM111Afamily with sequence similarity 111, member A (ENSG00000166801), score: 0.47 FAM119Bfamily with sequence similarity 119, member B (ENSG00000123427), score: 0.48 FAM151Afamily with sequence similarity 151, member A (ENSG00000162391), score: 0.61 FAM156Afamily with sequence similarity 156, member A (ENSG00000182646), score: -0.48 FAM55Cfamily with sequence similarity 55, member C (ENSG00000144815), score: -0.56 FAM83Hfamily with sequence similarity 83, member H (ENSG00000180921), score: 0.5 FAR1fatty acyl CoA reductase 1 (ENSG00000197601), score: -0.55 FBXO17F-box protein 17 (ENSG00000104835), score: 0.67 FCAMRFc receptor, IgA, IgM, high affinity (ENSG00000162897), score: 0.51 FCER1AFc fragment of IgE, high affinity I, receptor for; alpha polypeptide (ENSG00000179639), score: 0.49 FGFR4fibroblast growth factor receptor 4 (ENSG00000160867), score: 0.55 FRMD6FERM domain containing 6 (ENSG00000139926), score: -0.51 FTCDformiminotransferase cyclodeaminase (ENSG00000160282), score: 0.64 FUCA2fucosidase, alpha-L- 2, plasma (ENSG00000001036), score: 0.48 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (ENSG00000171124), score: 0.54 GAL3ST2galactose-3-O-sulfotransferase 2 (ENSG00000154252), score: 0.59 GALEUDP-galactose-4-epimerase (ENSG00000117308), score: 0.54 GANABglucosidase, alpha; neutral AB (ENSG00000089597), score: 0.59 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (ENSG00000171766), score: 0.47 GCDHglutaryl-CoA dehydrogenase (ENSG00000105607), score: 0.48 GDF7growth differentiation factor 7 (ENSG00000143869), score: 0.47 GGT6gamma-glutamyltransferase 6 (ENSG00000167741), score: 0.59 GHDCGH3 domain containing (ENSG00000167925), score: 0.51 GIPC2GIPC PDZ domain containing family, member 2 (ENSG00000137960), score: 0.51 GJC3gap junction protein, gamma 3, 30.2kDa (ENSG00000176402), score: 0.53 GLB1galactosidase, beta 1 (ENSG00000170266), score: 0.51 GLDCglycine dehydrogenase (decarboxylating) (ENSG00000178445), score: 0.47 GLTPD2glycolipid transfer protein domain containing 2 (ENSG00000182327), score: 0.66 GLYATL1glycine-N-acyltransferase-like 1 (ENSG00000166840), score: 0.49 GOSR2golgi SNAP receptor complex member 2 (ENSG00000108433), score: 0.48 GP1BAglycoprotein Ib (platelet), alpha polypeptide (ENSG00000185245), score: 0.6 GPD1glycerol-3-phosphate dehydrogenase 1 (soluble) (ENSG00000167588), score: 0.49 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 1 GRWD1glutamate-rich WD repeat containing 1 (ENSG00000105447), score: 0.52 HACE1HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 (ENSG00000085382), score: -0.49 HAO2hydroxyacid oxidase 2 (long chain) (ENSG00000116882), score: 0.46 HCN3hyperpolarization activated cyclic nucleotide-gated potassium channel 3 (ENSG00000143630), score: 0.53 HDAC6histone deacetylase 6 (ENSG00000094631), score: 0.61 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: 0.61 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: -0.61 HEXAhexosaminidase A (alpha polypeptide) (ENSG00000213614), score: 0.55 HHLA2HERV-H LTR-associating 2 (ENSG00000114455), score: 0.48 HINT2histidine triad nucleotide binding protein 2 (ENSG00000137133), score: 0.49 HMGCL3-hydroxymethyl-3-methylglutaryl-CoA lyase (ENSG00000117305), score: 0.51 HNF1AHNF1 homeobox A (ENSG00000135100), score: 0.54 HOMER1homer homolog 1 (Drosophila) (ENSG00000152413), score: -0.48 HPD4-hydroxyphenylpyruvate dioxygenase (ENSG00000158104), score: 0.5 HPNhepsin (ENSG00000105707), score: 0.47 HSCBHscB iron-sulfur cluster co-chaperone homolog (E. coli) (ENSG00000100209), score: 0.47 HSD17B14hydroxysteroid (17-beta) dehydrogenase 14 (ENSG00000087076), score: 0.53 HSPA1Lheat shock 70kDa protein 1-like (ENSG00000204390), score: -0.47 IL17RBinterleukin 17 receptor B (ENSG00000056736), score: 0.65 IL17RCinterleukin 17 receptor C (ENSG00000163702), score: 0.47 IL17REinterleukin 17 receptor E (ENSG00000163701), score: 0.6 IL1RL2interleukin 1 receptor-like 2 (ENSG00000115598), score: 0.57 IL22RA1interleukin 22 receptor, alpha 1 (ENSG00000142677), score: 0.64 IL9Rinterleukin 9 receptor (ENSG00000124334), score: 0.6 ILVBLilvB (bacterial acetolactate synthase)-like (ENSG00000105135), score: 0.47 INMTindolethylamine N-methyltransferase (ENSG00000106125), score: 0.49 INPP4Ainositol polyphosphate-4-phosphatase, type I, 107kDa (ENSG00000040933), score: -0.48 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.56 ISOC2isochorismatase domain containing 2 (ENSG00000063241), score: 0.47 JAGN1jagunal homolog 1 (Drosophila) (ENSG00000171135), score: 0.52 JAM2junctional adhesion molecule 2 (ENSG00000154721), score: -0.51 KBTBD2kelch repeat and BTB (POZ) domain containing 2 (ENSG00000170852), score: -0.5 KCNE3potassium voltage-gated channel, Isk-related family, member 3 (ENSG00000175538), score: 0.6 KCNH6potassium voltage-gated channel, subfamily H (eag-related), member 6 (ENSG00000173826), score: 0.61 KCNK5potassium channel, subfamily K, member 5 (ENSG00000164626), score: 0.55 KHKketohexokinase (fructokinase) (ENSG00000138030), score: 0.46 KIAA0391KIAA0391 (ENSG00000100890), score: 0.53 KIAA1191KIAA1191 (ENSG00000122203), score: 0.48 KLF7Kruppel-like factor 7 (ubiquitous) (ENSG00000118263), score: -0.56 KNG1kininogen 1 (ENSG00000113889), score: 0.48 KRT7keratin 7 (ENSG00000135480), score: 0.47 KRTCAP3keratinocyte associated protein 3 (ENSG00000157992), score: 0.48 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: 0.5 LASS2LAG1 homolog, ceramide synthase 2 (ENSG00000143418), score: 0.56 LCN12lipocalin 12 (ENSG00000184925), score: 0.55 LIPClipase, hepatic (ENSG00000166035), score: 0.55 LLGL2lethal giant larvae homolog 2 (Drosophila) (ENSG00000073350), score: 0.55 LOC55908hepatocellular carcinoma-associated gene TD26 (ENSG00000130173), score: 0.48 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: -0.54 LRRC8Eleucine rich repeat containing 8 family, member E (ENSG00000171017), score: 0.46 LYG1lysozyme G-like 1 (ENSG00000144214), score: 0.53 LYPD2LY6/PLAUR domain containing 2 (ENSG00000197353), score: 0.64 MAPKAPK5mitogen-activated protein kinase-activated protein kinase 5 (ENSG00000089022), score: 0.52 MCATmalonyl CoA:ACP acyltransferase (mitochondrial) (ENSG00000100294), score: 0.54 MCCD1mitochondrial coiled-coil domain 1 (ENSG00000204511), score: 0.52 METTL1methyltransferase like 1 (ENSG00000037897), score: 0.51 METTL6methyltransferase like 6 (ENSG00000206562), score: -0.48 MIFmacrophage migration inhibitory factor (glycosylation-inhibiting factor) (ENSG00000133460), score: 0.6 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.48 MLECmalectin (ENSG00000110917), score: 0.49 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.6 MOGAT1monoacylglycerol O-acyltransferase 1 (ENSG00000124003), score: 0.5 MPDU1mannose-P-dolichol utilization defect 1 (ENSG00000129255), score: 0.48 MRP63mitochondrial ribosomal protein 63 (ENSG00000173141), score: 0.47 MRPL49mitochondrial ribosomal protein L49 (ENSG00000149792), score: 0.5 MRPS18Bmitochondrial ribosomal protein S18B (ENSG00000204568), score: 0.49 MSH2mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) (ENSG00000095002), score: -0.54 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.49 MTHFS5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) (ENSG00000136371), score: 0.48 MUC13mucin 13, cell surface associated (ENSG00000173702), score: 0.55 MUC20mucin 20, cell surface associated (ENSG00000176945), score: 0.52 MVKmevalonate kinase (ENSG00000110921), score: 0.52 MYL5myosin, light chain 5, regulatory (ENSG00000215375), score: 0.55 MYST4MYST histone acetyltransferase (monocytic leukemia) 4 (ENSG00000156650), score: -0.52 NAGLUN-acetylglucosaminidase, alpha (ENSG00000108784), score: 0.54 NAGSN-acetylglutamate synthase (ENSG00000161653), score: 0.48 NCCRP1non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) (ENSG00000188505), score: 0.59 NDRG4NDRG family member 4 (ENSG00000103034), score: -0.51 NINJ1ninjurin 1 (ENSG00000131669), score: 0.53 NIT2nitrilase family, member 2 (ENSG00000114021), score: 0.52 NPBneuropeptide B (ENSG00000183979), score: 0.61 OAFOAF homolog (Drosophila) (ENSG00000184232), score: 0.46 OCIAD2OCIA domain containing 2 (ENSG00000145247), score: 0.46 ODF2Louter dense fiber of sperm tails 2-like (ENSG00000122417), score: -0.49 OS9osteosarcoma amplified 9, endoplasmic reticulum lectin (ENSG00000135506), score: 0.47 OSBPoxysterol binding protein (ENSG00000110048), score: 0.46 OXER1oxoeicosanoid (OXE) receptor 1 (ENSG00000162881), score: 0.57 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.7 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.52 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.46 PARK7Parkinson disease (autosomal recessive, early onset) 7 (ENSG00000116288), score: 0.47 PCDHGA12protocadherin gamma subfamily A, 12 (ENSG00000081853), score: -0.49 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.49 PDE4Bphosphodiesterase 4B, cAMP-specific (ENSG00000184588), score: -0.49 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.63 PELI1pellino homolog 1 (Drosophila) (ENSG00000197329), score: -0.54 PEPDpeptidase D (ENSG00000124299), score: 0.6 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: 0.49 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.51 PGAM5phosphoglycerate mutase family member 5 (ENSG00000176894), score: 0.64 PGAP3post-GPI attachment to proteins 3 (ENSG00000161395), score: 0.47 PGLS6-phosphogluconolactonase (ENSG00000130313), score: 0.52 PKLRpyruvate kinase, liver and RBC (ENSG00000143627), score: 0.62 PLA2G12Bphospholipase A2, group XIIB (ENSG00000138308), score: 0.5 PLEK2pleckstrin 2 (ENSG00000100558), score: 0.58 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.62 PLXNB2plexin B2 (ENSG00000196576), score: 0.49 PMVKphosphomevalonate kinase (ENSG00000163344), score: 0.53 PNPLA4patatin-like phospholipase domain containing 4 (ENSG00000006757), score: 0.52 PNPOpyridoxamine 5'-phosphate oxidase (ENSG00000108439), score: 0.48 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.6 PPP1R12Aprotein phosphatase 1, regulatory (inhibitor) subunit 12A (ENSG00000058272), score: -0.49 PRAP1proline-rich acidic protein 1 (ENSG00000165828), score: 0.55 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: 0.47 PRDM2PR domain containing 2, with ZNF domain (ENSG00000116731), score: -0.5 PRDX5peroxiredoxin 5 (ENSG00000126432), score: 0.46 PRICKLE4prickle homolog 4 (Drosophila) (ENSG00000124593), score: 0.52 PRKCSHprotein kinase C substrate 80K-H (ENSG00000130175), score: 0.5 PRLRprolactin receptor (ENSG00000113494), score: 0.55 PROCprotein C (inactivator of coagulation factors Va and VIIIa) (ENSG00000115718), score: 0.48 PRR13proline rich 13 (ENSG00000205352), score: 0.5 PRRG4proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) (ENSG00000135378), score: 0.61 PRSS8protease, serine, 8 (ENSG00000052344), score: 0.65 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.57 PSME2proteasome (prosome, macropain) activator subunit 2 (PA28 beta) (ENSG00000100911), score: 0.49 PTH1Rparathyroid hormone 1 receptor (ENSG00000160801), score: 0.5 PTPREprotein tyrosine phosphatase, receptor type, E (ENSG00000132334), score: -0.51 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: 0.54 RAB17RAB17, member RAS oncogene family (ENSG00000124839), score: 0.61 RAB8ARAB8A, member RAS oncogene family (ENSG00000167461), score: 0.55 RAG1AP1recombination activating gene 1 activating protein 1 (ENSG00000169241), score: 0.52 RALGAPA2Ral GTPase activating protein, alpha subunit 2 (catalytic) (ENSG00000188559), score: 0.57 RB1CC1RB1-inducible coiled-coil 1 (ENSG00000023287), score: -0.48 RBM26RNA binding motif protein 26 (ENSG00000139746), score: -0.5 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.63 RBPJrecombination signal binding protein for immunoglobulin kappa J region (ENSG00000168214), score: -0.5 RENrenin (ENSG00000143839), score: 0.47 RER1RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) (ENSG00000157916), score: 0.54 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (ENSG00000169733), score: 0.51 RHBGRh family, B glycoprotein (gene/pseudogene) (ENSG00000132677), score: 0.48 RIPPLY1ripply1 homolog (zebrafish) (ENSG00000147223), score: 0.5 RMND1required for meiotic nuclear division 1 homolog (S. cerevisiae) (ENSG00000155906), score: 0.51 RNF144Aring finger protein 144A (ENSG00000151692), score: -0.49 RNF145ring finger protein 145 (ENSG00000145860), score: -0.49 RNF34ring finger protein 34 (ENSG00000170633), score: -0.49 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: 0.62 ROBO1roundabout, axon guidance receptor, homolog 1 (Drosophila) (ENSG00000169855), score: -0.53 RUFY1RUN and FYVE domain containing 1 (ENSG00000176783), score: 0.51 SCRN2secernin 2 (ENSG00000141295), score: 0.52 SCUBE1signal peptide, CUB domain, EGF-like 1 (ENSG00000159307), score: 0.49 SDC1syndecan 1 (ENSG00000115884), score: 0.46 SEC61A1Sec61 alpha 1 subunit (S. cerevisiae) (ENSG00000058262), score: 0.47 SEPT7septin 7 (ENSG00000122545), score: -0.51 SERINC2serine incorporator 2 (ENSG00000168528), score: 0.46 SERPINA4serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 (ENSG00000100665), score: 0.49 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: -0.61 SFXN2sideroflexin 2 (ENSG00000156398), score: 0.46 SH3BP2SH3-domain binding protein 2 (ENSG00000087266), score: 0.52 SHHsonic hedgehog (ENSG00000164690), score: 0.65 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (ENSG00000185187), score: 0.51 SIVA1SIVA1, apoptosis-inducing factor (ENSG00000184990), score: 0.49 SKAP1src kinase associated phosphoprotein 1 (ENSG00000141293), score: 0.58 SLA2Src-like-adaptor 2 (ENSG00000101082), score: 0.61 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (ENSG00000088386), score: 0.78 SLC16A13solute carrier family 16, member 13 (monocarboxylic acid transporter 13) (ENSG00000174327), score: 0.62 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.69 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.78 SLC22A13solute carrier family 22 (organic anion transporter), member 13 (ENSG00000172940), score: 0.51 SLC22A7solute carrier family 22 (organic anion transporter), member 7 (ENSG00000137204), score: 0.51 SLC23A1solute carrier family 23 (nucleobase transporters), member 1 (ENSG00000170482), score: 0.51 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.58 SLC25A42solute carrier family 25, member 42 (ENSG00000181035), score: 0.64 SLC28A1solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 (ENSG00000156222), score: 0.55 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.54 SLC37A4solute carrier family 37 (glucose-6-phosphate transporter), member 4 (ENSG00000137700), score: 0.53 SLC39A4solute carrier family 39 (zinc transporter), member 4 (ENSG00000147804), score: 0.53 SLC39A5solute carrier family 39 (metal ion transporter), member 5 (ENSG00000139540), score: 0.78 SLC47A1solute carrier family 47, member 1 (ENSG00000142494), score: 0.57 SLC5A10solute carrier family 5 (sodium/glucose cotransporter), member 10 (ENSG00000154025), score: 0.51 SLC5A2solute carrier family 5 (sodium/glucose cotransporter), member 2 (ENSG00000140675), score: 0.51 SLC5A9solute carrier family 5 (sodium/glucose cotransporter), member 9 (ENSG00000117834), score: 0.53 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 0.51 SLCO2B1solute carrier organic anion transporter family, member 2B1 (ENSG00000137491), score: 0.52 SMOsmoothened homolog (Drosophila) (ENSG00000128602), score: 0.53 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (ENSG00000166311), score: 0.52 SMPDL3Bsphingomyelin phosphodiesterase, acid-like 3B (ENSG00000130768), score: 0.5 SNX22sorting nexin 22 (ENSG00000157734), score: 0.61 SP5Sp5 transcription factor (ENSG00000204335), score: 0.47 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (ENSG00000090487), score: 0.51 SRD5A2steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) (ENSG00000049319), score: 0.47 SSBP2single-stranded DNA binding protein 2 (ENSG00000145687), score: -0.5 SSR2signal sequence receptor, beta (translocon-associated protein beta) (ENSG00000163479), score: 0.47 ST13suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) (ENSG00000100380), score: 0.65 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (ENSG00000124214), score: 0.51 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.5 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: 0.51 STXBP5syntaxin binding protein 5 (tomosyn) (ENSG00000164506), score: -0.54 SUCLG1succinate-CoA ligase, alpha subunit (ENSG00000163541), score: 0.49 SUOXsulfite oxidase (ENSG00000139531), score: 0.53 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.48 TACSTD2tumor-associated calcium signal transducer 2 (ENSG00000184292), score: 0.48 TALDO1transaldolase 1 (ENSG00000177156), score: 0.52 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: 0.49 TBL1Xtransducin (beta)-like 1X-linked (ENSG00000101849), score: 0.52 TBX15T-box 15 (ENSG00000092607), score: 0.47 TCERG1transcription elongation regulator 1 (ENSG00000113649), score: -0.54 TCF4transcription factor 4 (ENSG00000196628), score: -0.57 TDGF1teratocarcinoma-derived growth factor 1 (ENSG00000241186), score: 0.52 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.75 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.57 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.53 TM7SF3transmembrane 7 superfamily member 3 (ENSG00000064115), score: 0.59 TMBIM6transmembrane BAX inhibitor motif containing 6 (ENSG00000139644), score: 0.47 TMCO4transmembrane and coiled-coil domains 4 (ENSG00000162542), score: 0.5 TMED4transmembrane emp24 protein transport domain containing 4 (ENSG00000158604), score: 0.61 TMEM115transmembrane protein 115 (ENSG00000126062), score: 0.56 TMEM139transmembrane protein 139 (ENSG00000178826), score: 0.47 TMEM150Btransmembrane protein 150B (ENSG00000180061), score: 0.66 TMEM161Atransmembrane protein 161A (ENSG00000064545), score: 0.47 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.47 TMEM177transmembrane protein 177 (ENSG00000144120), score: 0.57 TMEM195transmembrane protein 195 (ENSG00000187546), score: 0.5 TMEM208transmembrane protein 208 (ENSG00000168701), score: 0.49 TMEM219transmembrane protein 219 (ENSG00000149932), score: 0.47 TMEM37transmembrane protein 37 (ENSG00000171227), score: 0.5 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.76 TMEM82transmembrane protein 82 (ENSG00000162460), score: 0.5 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.47 TNRC6Ctrinucleotide repeat containing 6C (ENSG00000078687), score: -0.52 TOMM22translocase of outer mitochondrial membrane 22 homolog (yeast) (ENSG00000100216), score: 0.52 TOR3Atorsin family 3, member A (ENSG00000186283), score: 0.52 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.52 TRAP1TNF receptor-associated protein 1 (ENSG00000126602), score: 0.57 TRIM10tripartite motif-containing 10 (ENSG00000204613), score: 0.84 TRIM15tripartite motif-containing 15 (ENSG00000204610), score: 0.93 TRIM47tripartite motif-containing 47 (ENSG00000132481), score: 0.48 TRIP6thyroid hormone receptor interactor 6 (ENSG00000087077), score: 0.68 TRPC7transient receptor potential cation channel, subfamily C, member 7 (ENSG00000069018), score: 0.67 TSPAN16tetraspanin 16 (ENSG00000130167), score: 0.53 TSPAN33tetraspanin 33 (ENSG00000158457), score: 0.57 TSPO2translocator protein 2 (ENSG00000112212), score: 0.59 TSTD1thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 (ENSG00000215845), score: 0.48 TTC22tetratricopeptide repeat domain 22 (ENSG00000006555), score: 0.54 TUBAL3tubulin, alpha-like 3 (ENSG00000178462), score: 0.64 TXNRD2thioredoxin reductase 2 (ENSG00000184470), score: 0.5 UGT2B7UDP glucuronosyltransferase 2 family, polypeptide B7 (ENSG00000171234), score: 0.56 UMPSuridine monophosphate synthetase (ENSG00000114491), score: 0.51 UNC5CLunc-5 homolog C (C. elegans)-like (ENSG00000124602), score: 0.51 USH2AUsher syndrome 2A (autosomal recessive, mild) (ENSG00000042781), score: 0.61 UTS2urotensin 2 (ENSG00000049247), score: 0.52 VAV2vav 2 guanine nucleotide exchange factor (ENSG00000160293), score: 0.57 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.51 WASF3WAS protein family, member 3 (ENSG00000132970), score: -0.52 WDR18WD repeat domain 18 (ENSG00000065268), score: 0.48 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.55 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.52 YEATS2YEATS domain containing 2 (ENSG00000163872), score: -0.49 ZBTB46zinc finger and BTB domain containing 46 (ENSG00000130584), score: 0.5 ZC3H6zinc finger CCCH-type containing 6 (ENSG00000188177), score: -0.59 ZEB1zinc finger E-box binding homeobox 1 (ENSG00000148516), score: -0.49 ZFP28zinc finger protein 28 homolog (mouse) (ENSG00000196867), score: -0.49 ZG16zymogen granule protein 16 homolog (rat) (ENSG00000174992), score: 0.66 ZGPATzinc finger, CCCH-type with G patch domain (ENSG00000197114), score: 0.59 ZNF192zinc finger protein 192 (ENSG00000198315), score: -0.48 ZNF248zinc finger protein 248 (ENSG00000198105), score: -0.48 ZNF292zinc finger protein 292 (ENSG00000188994), score: -0.51 ZNF511zinc finger protein 511 (ENSG00000198546), score: 0.49 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.62 ZNF599zinc finger protein 599 (ENSG00000153896), score: 0.54 ZSCAN2zinc finger and SCAN domain containing 2 (ENSG00000176371), score: 0.48
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |