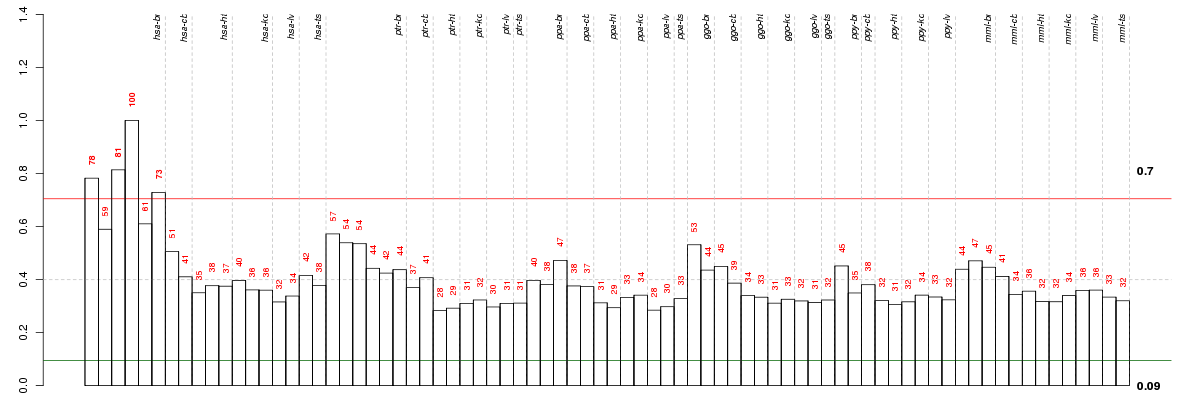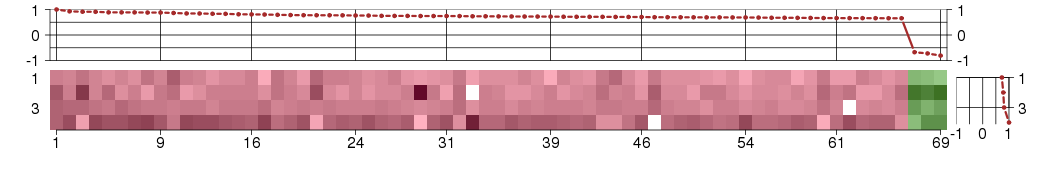

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
ATP10BATPase, class V, type 10B (ENSG00000118322), score: 0.71 C11orf9chromosome 11 open reading frame 9 (ENSG00000124920), score: 0.69 C1orf198chromosome 1 open reading frame 198 (ENSG00000119280), score: 0.72 C3orf16chromosome 3 open reading frame 16 (ENSG00000206199), score: 0.71 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: -0.81 CD22CD22 molecule (ENSG00000012124), score: 0.91 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.88 CHADLchondroadherin-like (ENSG00000100399), score: 0.81 CLCA4chloride channel accessory 4 (ENSG00000016602), score: 0.85 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.72 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.89 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.86 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (ENSG00000173786), score: 0.76 CPNE2copine II (ENSG00000140848), score: 0.67 CRYBB1crystallin, beta B1 (ENSG00000100122), score: 0.71 CYB5D1cytochrome b5 domain containing 1 (ENSG00000182224), score: -0.72 DEFB134defensin, beta 134 (ENSG00000205882), score: 0.77 DMBT1deleted in malignant brain tumors 1 (ENSG00000187908), score: 0.8 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.78 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.78 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.68 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.66 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.68 FABP6fatty acid binding protein 6, ileal (ENSG00000170231), score: 0.74 FAM108B1family with sequence similarity 108, member B1 (ENSG00000107362), score: 0.66 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.83 FAM125Bfamily with sequence similarity 125, member B (ENSG00000196814), score: 0.71 GAB2GRB2-associated binding protein 2 (ENSG00000033327), score: 0.67 GABRQgamma-aminobutyric acid (GABA) receptor, theta (ENSG00000147402), score: 0.7 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (ENSG00000139629), score: 0.89 GIPRgastric inhibitory polypeptide receptor (ENSG00000010310), score: 0.73 GPR78G protein-coupled receptor 78 (ENSG00000155269), score: 0.74 GREM1gremlin 1 (ENSG00000166923), score: 0.88 GTF2E2general transcription factor IIE, polypeptide 2, beta 34kDa (ENSG00000197265), score: -0.67 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.66 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.7 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.75 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.73 KREMEN2kringle containing transmembrane protein 2 (ENSG00000131650), score: 1 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.84 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (ENSG00000107902), score: 0.74 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.79 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.91 MARCKSL1MARCKS-like 1 (ENSG00000175130), score: 0.73 MYO9Bmyosin IXB (ENSG00000099331), score: 0.69 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.89 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.93 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.72 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.73 PRKCQprotein kinase C, theta (ENSG00000065675), score: 0.68 PRR18proline rich 18 (ENSG00000176381), score: 0.81 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.66 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.77 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.69 PTPRHprotein tyrosine phosphatase, receptor type, H (ENSG00000080031), score: 0.83 RAB4BRAB4B, member RAS oncogene family (ENSG00000167578), score: 0.71 RHOGras homolog gene family, member G (rho G) (ENSG00000177105), score: 0.72 ROM1retinal outer segment membrane protein 1 (ENSG00000149489), score: 0.69 SEMA4Dsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D (ENSG00000187764), score: 0.69 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.69 SH3TC2SH3 domain and tetratricopeptide repeats 2 (ENSG00000169247), score: 0.74 SIRT2sirtuin 2 (ENSG00000068903), score: 0.76 SYT15synaptotagmin XV (ENSG00000204176), score: 0.66 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.75 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.77 TNFRSF13Ctumor necrosis factor receptor superfamily, member 13C (ENSG00000159958), score: 0.66 TP53INP2tumor protein p53 inducible nuclear protein 2 (ENSG00000078804), score: 0.71 UNC5Cunc-5 homolog C (C. elegans) (ENSG00000182168), score: 0.75 ZSWIM6zinc finger, SWIM-type containing 6 (ENSG00000130449), score: 0.66
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |