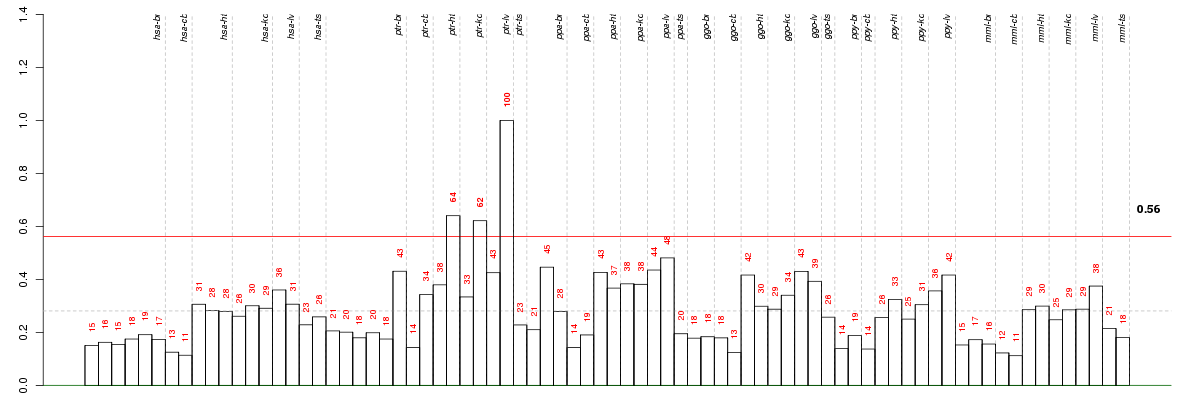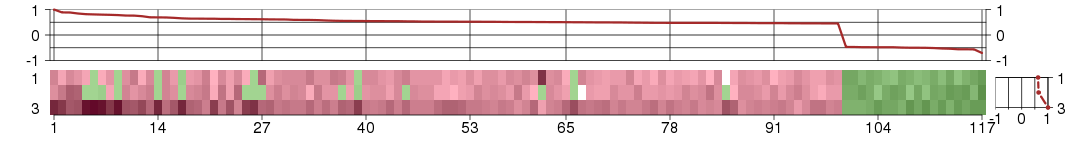

Under-expression is coded with green,
over-expression with red color.

MAPKKK cascade
A cascade of at least three protein kinase activities culminating in the phosphorylation and activation of a MAP kinase. MAPKKK cascades lie downstream of numerous signaling pathways.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
activation of innate immune response
Any process that initiates an innate immune response. Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. Examples of this process include activation of the hypersensitive response of Arabidopsis thaliana and activation of any NOD or TLR signaling pathway in vertebrate species.
pattern recognition receptor signaling pathway
Any series of molecular signals generated as a consequence of binding to a cell surface or intracellular pattern recognition receptor (PRR). Such receptors bind for molecular patterns based on a repeating or polymeric structures, like those of polysaccharides or peptidoglycans, which are sometimes associated with potential pathogens.
toll-like receptor signaling pathway
Any series of molecular signals generated as a consequence of binding to a toll-like receptor. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate innate immune response.
response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.
adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies). An example of this is the adaptive immune response found in Mus musculus.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune response-activating signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an immune response.
innate immune response-activating signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an innate immune response.
immune response-regulating signaling pathway
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to the activation, perpetuation, or inhibition of an immune response.
regulation of adaptive immune response
Any process that modulates the frequency, rate, or extent of an adaptive immune response.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that modulates the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains. An example of this process is found in the Gnathostomata.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains. An example of this process is found in the Gnathostomata.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to virus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
intracellular protein kinase cascade
A series of reactions that occur within the cell, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
JNK cascade
A cascade of protein kinase activities, culminating in the phosphorylation and activation of a member of the JUN kinase subfamily of stress-activated protein kinases, which in turn are a subfamily of mitogen-activated protein (MAP) kinases that is activated primarily by cytokines and exposure to environmental stress.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
response to bacterium
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to organic substance
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic substance stimulus.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of intracellular protein kinase cascade
Any process that modulates the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
positive regulation of intracellular protein kinase cascade
Any process that increases the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cytokine-mediated signaling pathway
Any series of molecular signals generated as a consequence of a cytokine or chemokine binding to a cell surface receptor.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
signal transmission via phosphorylation event
The process whereby a signal is conveyed via the transfer of one or more phosphate groups.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
initiation of signal transduction
The process whereby a signal causes activation of a receptor, for example, via a conformation change.
signal initiation by diffusible mediator
The process whereby a diffusible signal causes activation of a receptor.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signal initiation by protein/peptide mediator
The process whereby a protein/peptide signal causes activation of a receptor.
regulation of signaling process
Any process that modulates the frequency, rate or extent of a signaling process.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
positive regulation of signaling process
Any process that activates, maintains or increases the frequency, rate or extent of a signaling process.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
stress-activated protein kinase signaling cascade
A series of molecular signals in which a stress-activated protein kinase (SAPK) cascade relays one or more of the signals.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of defense response
Any process that modulates the frequency, rate or extent of a defense response.
positive regulation of defense response
Any process that activates or increases the frequency, rate or extent of a defense response.
negative regulation of NF-kappaB transcription factor activity
Any process that stops, prevents or reduces the frequency, rate or extent of the activity of the transcription factor NF-kappaB.
response to peptidoglycan
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptidoglycan stimulus. Peptidoglycan is a bacterial cell wall macromolecule.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
toll-like receptor 2 signaling pathway
Any series of molecular signals generated as a consequence of binding to toll-like receptor 2.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of signaling pathway
Any process that modulates the frequency, rate or extent of a signaling pathway.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
response to exogenous dsRNA
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an exogenous double-stranded RNA stimulus.
response to dsRNA
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a double-stranded RNA stimulus.
positive regulation of DNA binding
Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
negative regulation of DNA binding
Any process that stops or reduces the frequency, rate or extent of DNA binding. DNA binding is any process by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
regulation of MAPKKK cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAPKKK cascade.
positive regulation of MAPKKK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade.
negative regulation of transcription factor activity
Any process that stops, prevents or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
negative regulation of molecular function
Any process that stops or reduces the rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
positive regulation of molecular function
Any process that activates or increases the rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
regulation of innate immune response
Any process that modulates the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
positive regulation of innate immune response
Any process that activates or increases the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of JNK cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the JNK cascade.
positive regulation of JNK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of transcription factor activity
Any process that modulates the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
positive regulation of transcription factor activity
Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
positive regulation of NF-kappaB transcription factor activity
Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB.
regulation of binding
Any process that modulates the frequency, rate or extent of binding, the selective interaction of a molecule with one or more specific sites on another molecule.
positive regulation of binding
Any process that activates or increases the rate or extent of binding, the selective interaction of a molecule with one or more specific sites on another molecule.
negative regulation of binding
Any process that stops or reduces the rate or extent of binding, the selective interaction of a molecule with one or more specific sites on another molecule.
regulation of DNA binding
Any process that modulates the frequency, rate or extent of DNA binding. DNA binding is any process by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
regulation of stress-activated protein kinase signaling cascade
Any process that modulates the frequency, rate or extent of signaling via a stress-activated protein kinase signaling cascade.
positive regulation of stress-activated protein kinase signaling cascade
Any process that activates or increases the frequency, rate or extent of signaling via the stress-activated protein kinase signaling cascade.
ERK1 and ERK2 cascade
A cascade of protein kinase activities, culminating in the phosphorylation and activation of either the ERK1 or ERK2 kinases, which in turn are a subfamily of mitogen-activated protein (MAP) kinases.
regulation of ERK1 and ERK2 cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade.
positive regulation of ERK1 and ERK2 cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of cellular response to stress
Any process that modulates the frequency, rate or extent of a cellular response to stress. Cellular response to stress is a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of transcription regulator activity
Any process that modulates the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
positive regulation of transcription regulator activity
Any process that increases the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
negative regulation of transcription regulator activity
Any process that decreases the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of signaling process
Any process that modulates the frequency, rate or extent of a signaling process.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
regulation of signaling pathway
Any process that modulates the frequency, rate or extent of a signaling pathway.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
regulation of response to stress
Any process that modulates the frequency, rate or extent of a response to stress. Response to stress is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
regulation of transcription regulator activity
Any process that modulates the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
innate immune response-activating signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an innate immune response.
immune response-regulating signaling pathway
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to the activation, perpetuation, or inhibition of an immune response.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
regulation of adaptive immune response
Any process that modulates the frequency, rate, or extent of an adaptive immune response.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
regulation of innate immune response
Any process that modulates the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
positive regulation of innate immune response
Any process that activates or increases the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
initiation of signal transduction
The process whereby a signal causes activation of a receptor, for example, via a conformation change.
regulation of cellular response to stress
Any process that modulates the frequency, rate or extent of a cellular response to stress. Cellular response to stress is a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
regulation of intracellular protein kinase cascade
Any process that modulates the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
immune response-activating signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to activation or perpetuation of an immune response.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
positive regulation of innate immune response
Any process that activates or increases the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
positive regulation of intracellular protein kinase cascade
Any process that increases the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
regulation of stress-activated protein kinase signaling cascade
Any process that modulates the frequency, rate or extent of signaling via a stress-activated protein kinase signaling cascade.
regulation of cellular response to stress
Any process that modulates the frequency, rate or extent of a cellular response to stress. Cellular response to stress is a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
positive regulation of transcription factor activity
Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
negative regulation of transcription factor activity
Any process that stops, prevents or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
regulation of defense response
Any process that modulates the frequency, rate or extent of a defense response.
positive regulation of defense response
Any process that activates or increases the frequency, rate or extent of a defense response.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.
response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.
negative regulation of transcription regulator activity
Any process that decreases the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
positive regulation of transcription regulator activity
Any process that increases the frequency, rate or extent of transcription regulator activity, any molecular function that plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
positive regulation of binding
Any process that activates or increases the rate or extent of binding, the selective interaction of a molecule with one or more specific sites on another molecule.
negative regulation of binding
Any process that stops or reduces the rate or extent of binding, the selective interaction of a molecule with one or more specific sites on another molecule.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
positive regulation of adaptive immune response
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response.
positive regulation of innate immune response
Any process that activates or increases the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
activation of innate immune response
Any process that initiates an innate immune response. Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. Examples of this process include activation of the hypersensitive response of Arabidopsis thaliana and activation of any NOD or TLR signaling pathway in vertebrate species.
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that modulates the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains. An example of this process is found in the Gnathostomata.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains. An example of this process is found in the Gnathostomata.
positive regulation of intracellular protein kinase cascade
Any process that increases the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
intracellular protein kinase cascade
A series of reactions that occur within the cell, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
positive regulation of stress-activated protein kinase signaling cascade
Any process that activates or increases the frequency, rate or extent of signaling via the stress-activated protein kinase signaling cascade.
regulation of stress-activated protein kinase signaling cascade
Any process that modulates the frequency, rate or extent of signaling via a stress-activated protein kinase signaling cascade.
positive regulation of stress-activated protein kinase signaling cascade
Any process that activates or increases the frequency, rate or extent of signaling via the stress-activated protein kinase signaling cascade.
cytokine-mediated signaling pathway
Any series of molecular signals generated as a consequence of a cytokine or chemokine binding to a cell surface receptor.
regulation of intracellular protein kinase cascade
Any process that modulates the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
positive regulation of intracellular protein kinase cascade
Any process that increases the rate, frequency or extent of a series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
stress-activated protein kinase signaling cascade
A series of molecular signals in which a stress-activated protein kinase (SAPK) cascade relays one or more of the signals.
positive regulation of stress-activated protein kinase signaling cascade
Any process that activates or increases the frequency, rate or extent of signaling via the stress-activated protein kinase signaling cascade.
positive regulation of MAPKKK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade.
regulation of JNK cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the JNK cascade.
positive regulation of defense response
Any process that activates or increases the frequency, rate or extent of a defense response.
regulation of innate immune response
Any process that modulates the frequency, rate or extent of the innate immune response, the organism's first line of defense against infection.
positive regulation of DNA binding
Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
negative regulation of DNA binding
Any process that stops or reduces the frequency, rate or extent of DNA binding. DNA binding is any process by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
regulation of transcription factor activity
Any process that modulates the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
regulation of transcription factor activity
Any process that modulates the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
Any process that activates or increases the frequency, rate, or extent of an adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains. An example of this process is found in the Gnathostomata.
positive regulation of JNK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade.
regulation of JNK cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the JNK cascade.
positive regulation of JNK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade.
JNK cascade
A cascade of protein kinase activities, culminating in the phosphorylation and activation of a member of the JUN kinase subfamily of stress-activated protein kinases, which in turn are a subfamily of mitogen-activated protein (MAP) kinases that is activated primarily by cytokines and exposure to environmental stress.
regulation of MAPKKK cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAPKKK cascade.
positive regulation of MAPKKK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade.
positive regulation of JNK cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade.
positive regulation of ERK1 and ERK2 cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade.
negative regulation of transcription factor activity
Any process that stops, prevents or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
positive regulation of transcription factor activity
Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription.
regulation of ERK1 and ERK2 cascade
Any process that modulates the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade.
positive regulation of ERK1 and ERK2 cascade
Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade.


nucleotide binding
Interacting selectively and non-covalently with a nucleotide, any compound consisting of a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the ribose or deoxyribose moiety.
protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
G-protein-coupled receptor binding
Interacting selectively and non-covalently with a G-protein-coupled receptor.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively and non-covalently with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
cytokine receptor binding
Interacting selectively with a cytokine receptor.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
chemokine activity
The function of a family of chemotactic pro-inflammatory activation-inducible cytokines acting primarily upon hemopoietic cells in immunoregulatory processes; all chemokines possess a number of conserved cysteine residues involved in intramolecular disulfide bond formation.
purine nucleotide binding
Interacting selectively and non-covalently with purine nucleotides, any compound consisting of a purine nucleoside esterified with (ortho)phosphate.
chemokine receptor binding
Interacting selectively and non-covalently with any chemokine receptor.
all
NA
chemokine receptor binding
Interacting selectively and non-covalently with any chemokine receptor.
chemokine activity
The function of a family of chemotactic pro-inflammatory activation-inducible cytokines acting primarily upon hemopoietic cells in immunoregulatory processes; all chemokines possess a number of conserved cysteine residues involved in intramolecular disulfide bond formation.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04620 | 4.204e-04 | 0.7925 | 7 | 76 | Toll-like receptor signaling pathway |
| 04622 | 4.605e-04 | 0.5318 | 6 | 51 | RIG-I-like receptor signaling pathway |
| 05310 | 2.435e-02 | 0.219 | 3 | 21 | Asthma |
| 04621 | 2.719e-02 | 0.5005 | 4 | 48 | NOD-like receptor signaling pathway |
| 05140 | 3.216e-02 | 0.5318 | 4 | 51 | Leishmaniasis |
| 05322 | 3.421e-02 | 0.5422 | 4 | 52 | Systemic lupus erythematosus |
| 04062 | 4.976e-02 | 1.47 | 6 | 141 | Chemokine signaling pathway |
ADAMDEC1ADAM-like, decysin 1 (ENSG00000134028), score: 0.55 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.82 BATF2basic leucine zipper transcription factor, ATF-like 2 (ENSG00000168062), score: 0.59 C22orf28chromosome 22 open reading frame 28 (ENSG00000100220), score: 0.51 CALCBcalcitonin-related polypeptide beta (ENSG00000175868), score: 0.54 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: -0.53 CCL20chemokine (C-C motif) ligand 20 (ENSG00000115009), score: 0.62 CD244CD244 molecule, natural killer cell receptor 2B4 (ENSG00000122223), score: 0.47 CD274CD274 molecule (ENSG00000120217), score: 0.47 CD2BP2CD2 (cytoplasmic tail) binding protein 2 (ENSG00000169217), score: 0.48 CDYLchromodomain protein, Y-like (ENSG00000153046), score: 0.48 CMPK2cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial (ENSG00000134326), score: 0.61 CRELD1cysteine-rich with EGF-like domains 1 (ENSG00000163703), score: -0.56 CTSFcathepsin F (ENSG00000174080), score: -0.47 CXCL10chemokine (C-X-C motif) ligand 10 (ENSG00000169245), score: 0.76 CXCL11chemokine (C-X-C motif) ligand 11 (ENSG00000169248), score: 0.84 CXCL9chemokine (C-X-C motif) ligand 9 (ENSG00000138755), score: 0.74 CXorf38chromosome X open reading frame 38 (ENSG00000185753), score: 0.49 CYBBcytochrome b-245, beta polypeptide (ENSG00000165168), score: 0.52 CYHR1cysteine/histidine-rich 1 (ENSG00000187954), score: -0.48 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (ENSG00000141141), score: 0.47 DDX58DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (ENSG00000107201), score: 0.59 DHPSdeoxyhypusine synthase (ENSG00000095059), score: -0.54 DKC1dyskeratosis congenita 1, dyskerin (ENSG00000130826), score: 0.48 DUOX2dual oxidase 2 (ENSG00000140279), score: 0.8 DUOXA2dual oxidase maturation factor 2 (ENSG00000140274), score: 0.81 EPSTI1epithelial stromal interaction 1 (breast) (ENSG00000133106), score: 0.69 ETV6ets variant 6 (ENSG00000139083), score: 0.49 ETV7ets variant 7 (ENSG00000010030), score: 0.76 FARSAphenylalanyl-tRNA synthetase, alpha subunit (ENSG00000179115), score: 0.46 FFAR2free fatty acid receptor 2 (ENSG00000126262), score: 0.79 FGF19fibroblast growth factor 19 (ENSG00000162344), score: 0.78 FKBP4FK506 binding protein 4, 59kDa (ENSG00000004478), score: -0.47 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: 0.46 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (ENSG00000117228), score: 0.54 GBP3guanylate binding protein 3 (ENSG00000117226), score: 0.47 GOLGA5golgin A5 (ENSG00000066455), score: 0.48 GPR82G protein-coupled receptor 82 (ENSG00000171657), score: 0.89 HIST2H2ABhistone cluster 2, H2ab (ENSG00000184270), score: 1 HLA-DOBmajor histocompatibility complex, class II, DO beta (ENSG00000204267), score: 0.48 HRCT1histidine rich carboxyl terminus 1 (ENSG00000196196), score: 0.51 ICAM1intercellular adhesion molecule 1 (ENSG00000090339), score: 0.55 IDO1indoleamine 2,3-dioxygenase 1 (ENSG00000131203), score: 0.51 IFI44interferon-induced protein 44 (ENSG00000137965), score: 0.53 IFIT1interferon-induced protein with tetratricopeptide repeats 1 (ENSG00000185745), score: 0.51 IFIT2interferon-induced protein with tetratricopeptide repeats 2 (ENSG00000119922), score: 0.58 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (ENSG00000119917), score: 0.69 IL17Finterleukin 17F (ENSG00000112116), score: 0.51 IL2RAinterleukin 2 receptor, alpha (ENSG00000134460), score: 0.88 IL8interleukin 8 (ENSG00000169429), score: 0.47 INTS3integrator complex subunit 3 (ENSG00000143624), score: -0.56 IRAK1interleukin-1 receptor-associated kinase 1 (ENSG00000184216), score: 0.51 IRF1interferon regulatory factor 1 (ENSG00000125347), score: 0.53 ISG15ISG15 ubiquitin-like modifier (ENSG00000187608), score: 0.52 ISG20interferon stimulated exonuclease gene 20kDa (ENSG00000172183), score: 0.54 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: -0.48 ITLN2intelectin 2 (ENSG00000158764), score: 0.62 KCTD10potassium channel tetramerisation domain containing 10 (ENSG00000110906), score: 0.46 KPTNkaptin (actin binding protein) (ENSG00000118162), score: -0.5 LAP3leucine aminopeptidase 3 (ENSG00000002549), score: 0.5 LIPKlipase, family member K (ENSG00000204021), score: 0.5 LYSMD1LysM, putative peptidoglycan-binding, domain containing 1 (ENSG00000163155), score: -0.5 MAP4K2mitogen-activated protein kinase kinase kinase kinase 2 (ENSG00000168067), score: -0.48 MBOAT4membrane bound O-acyltransferase domain containing 4 (ENSG00000177669), score: 0.65 MPEG1macrophage expressed 1 (ENSG00000197629), score: 0.51 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (ENSG00000157601), score: 0.61 NLRC5NLR family, CARD domain containing 5 (ENSG00000140853), score: 0.52 NOD2nucleotide-binding oligomerization domain containing 2 (ENSG00000167207), score: 0.64 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (ENSG00000089127), score: 0.54 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (ENSG00000111335), score: 0.48 OASL2'-5'-oligoadenylate synthetase-like (ENSG00000135114), score: 0.5 OPN1SWopsin 1 (cone pigments), short-wave-sensitive (ENSG00000128617), score: 0.46 PARLpresenilin associated, rhomboid-like (ENSG00000175193), score: 0.48 PARP14poly (ADP-ribose) polymerase family, member 14 (ENSG00000173193), score: 0.52 PARP9poly (ADP-ribose) polymerase family, member 9 (ENSG00000138496), score: 0.54 PDCD7programmed cell death 7 (ENSG00000090470), score: -0.49 PEX6peroxisomal biogenesis factor 6 (ENSG00000124587), score: -0.7 PRAMEF12PRAME family member 12 (ENSG00000116726), score: 0.69 PRG2proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) (ENSG00000186652), score: 0.63 PRIC285peroxisomal proliferator-activated receptor A interacting complex 285 (ENSG00000130589), score: 0.45 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (ENSG00000240065), score: 0.45 PSMD14proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 (ENSG00000115233), score: 0.48 RIPK2receptor-interacting serine-threonine kinase 2 (ENSG00000104312), score: 0.61 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.47 RSAD2radical S-adenosyl methionine domain containing 2 (ENSG00000134321), score: 0.63 RTP4receptor (chemosensory) transporter protein 4 (ENSG00000136514), score: 0.52 RUNX1runt-related transcription factor 1 (ENSG00000159216), score: 0.49 SAMD9sterile alpha motif domain containing 9 (ENSG00000205413), score: 0.62 SERPINB8serpin peptidase inhibitor, clade B (ovalbumin), member 8 (ENSG00000166401), score: 0.64 SGCEsarcoglycan, epsilon (ENSG00000127990), score: -0.5 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: 0.52 SLC35A5solute carrier family 35, member A5 (ENSG00000138459), score: -0.56 SLC35B1solute carrier family 35, member B1 (ENSG00000121073), score: 0.53 SLFN5schlafen family member 5 (ENSG00000166750), score: 0.48 SNX32sorting nexin 32 (ENSG00000172803), score: -0.48 SOD2superoxide dismutase 2, mitochondrial (ENSG00000112096), score: 0.55 SPC24SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000161888), score: 0.48 STAT1signal transducer and activator of transcription 1, 91kDa (ENSG00000115415), score: 0.64 STK19serine/threonine kinase 19 (ENSG00000204344), score: 0.59 TCOF1Treacher Collins-Franceschetti syndrome 1 (ENSG00000070814), score: 0.56 TFPI2tissue factor pathway inhibitor 2 (ENSG00000105825), score: 0.46 TMEM156transmembrane protein 156 (ENSG00000121895), score: 0.55 TNFtumor necrosis factor (ENSG00000232810), score: 0.52 TRAFD1TRAF-type zinc finger domain containing 1 (ENSG00000135148), score: 0.47 TRIM21tripartite motif-containing 21 (ENSG00000132109), score: 0.47 TRIM22tripartite motif-containing 22 (ENSG00000132274), score: 0.49 TRIM25tripartite motif-containing 25 (ENSG00000121060), score: 0.52 TYMPthymidine phosphorylase (ENSG00000025708), score: 0.5 UBE2L6ubiquitin-conjugating enzyme E2L 6 (ENSG00000156587), score: 0.49 USP21ubiquitin specific peptidase 21 (ENSG00000143258), score: -0.48 ZBP1Z-DNA binding protein 1 (ENSG00000124256), score: 0.67 ZC3H12Azinc finger CCCH-type containing 12A (ENSG00000163874), score: 0.5 ZFYVE19zinc finger, FYVE domain containing 19 (ENSG00000166140), score: -0.51 ZNF480zinc finger protein 480 (ENSG00000198464), score: 0.47 ZNFX1zinc finger, NFX1-type containing 1 (ENSG00000124201), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| ptr_lv_f_ca1 | ptr | lv | f | _ |