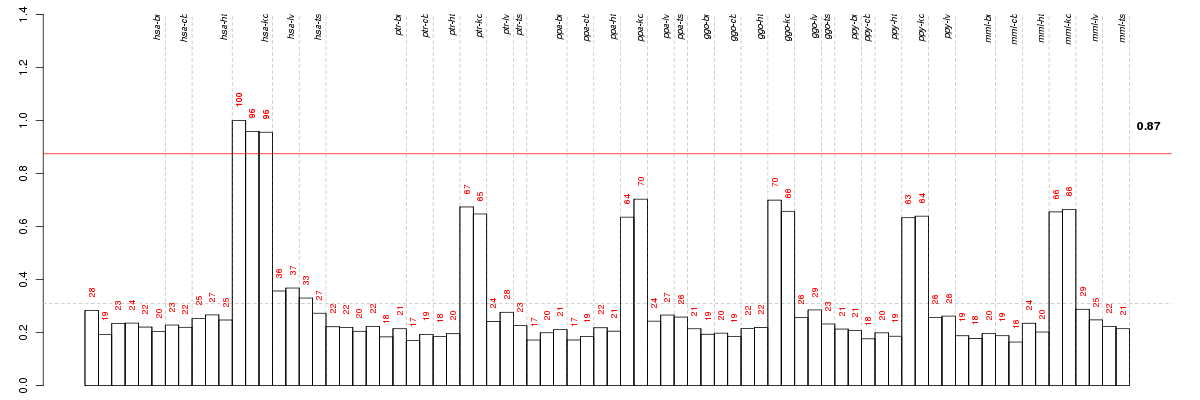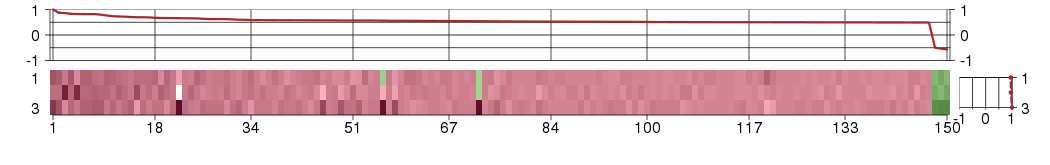

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
renal system process involved in regulation of blood volume
A slow mechanism of blood pressure regulation that responds to changes in pressure resulting from fluid and salt intake by modulating the quantity of blood in the circulatory system.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
circulatory system process
A organ system process carried out by any of the organs or tissues of the circulatory system. The circulatory system is an organ system that moves extracellular fluids to and from tissue within a multicellular organism.
renal system process
A organ system process carried out by any of the organs or tissues of the renal system. The renal system is responsible for fluid volume regulation and detoxification in an organism.
renal system process involved in regulation of systemic arterial blood pressure
Renal process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure.
regulation of systemic arterial blood pressure
The process that modulates the force with which blood travels through the systemic arterial circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure.
renal water transport
The directed movement of water (H2O) by the kidney.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
membrane lipid metabolic process
The chemical reactions and pathways involving membrane lipids, any lipid found in or associated with a biological membrane.
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
sphingolipid metabolic process
The chemical reactions and pathways involving sphingolipids, any of a class of lipids containing the long-chain amine diol sphingosine or a closely related base (a sphingoid).
sphingomyelin metabolic process
The chemical reactions and pathways involving sphingomyelin, N-acyl-4-sphingenyl-1-O-phosphorylcholine, any of a class of phospholipids in which the amino group of sphingosine is in amide linkage with one of several fatty acids, while the terminal hydroxyl group of sphingosine is esterified to phosphorylcholine.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
water transport
The directed movement of water (H2O) into, out of, within or between cells by means of some external agent such as a transporter or pore.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
blood circulation
The flow of blood through the body of an animal, enabling the transport of nutrients to the tissues and the removal of waste products.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
regulation of blood pressure
Any process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure.
carbohydrate transport
The directed movement of carbohydrate into, out of, within or between cells by means of some external agent such as a transporter or pore. Carbohydrates are any of a group of organic compounds based of the general formula Cx(H2O)y.
response to metal ion
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a metal ion stimulus.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to inorganic substance
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an inorganic substance stimulus.
monovalent inorganic cation transport
The directed movement of inorganic cations with a valency of one into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
polyol transport
The directed movement of polyols, any polyhydric alcohol, into, out of, within or between cells by means of some external agent such as a transporter or pore.
glycerol transport
The directed movement of glycerol into, out of, within or between cells by means of some external agent such as a transporter or pore. Glycerol is 1,2,3-propanetriol, a sweet, hygroscopic, viscous liquid, widely distributed in nature as a constituent of many lipids.
organic alcohol transport
The directed movement of organic alcohols into, out of, within or between cells by means of some external agent such as a transporter or pore. An organic alcohol is any carbon-containing compound containing a hydroxyl group.
organophosphate metabolic process
The chemical reactions and pathways involving organophosphates, any phosphate-containing organic compound.
response to mercury ion
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mercury ion stimulus.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
fluid transport
The directed movement of substances that are in liquid form in normal living conditions into, out of, within or between cells by means of some external agent such as a transporter or pore.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
response to copper ion
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a copper ion stimulus.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
cellular response to chemical stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
cellular response to inorganic substance
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an inorganic substance stimulus.
cellular response to metal ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a metal ion stimulus.
cellular response to copper ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a copper ion stimulus.
cellular response to mercury ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mercury ion stimulus.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular response to chemical stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
phospholipid metabolic process
The chemical reactions and pathways involving phospholipids, any lipid containing phosphoric acid as a mono- or diester.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
renal water transport
The directed movement of water (H2O) by the kidney.
cellular response to inorganic substance
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an inorganic substance stimulus.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
regulation of blood pressure
Any process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure.
renal system process involved in regulation of blood volume
A slow mechanism of blood pressure regulation that responds to changes in pressure resulting from fluid and salt intake by modulating the quantity of blood in the circulatory system.
cellular response to metal ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a metal ion stimulus.
glycerol transport
The directed movement of glycerol into, out of, within or between cells by means of some external agent such as a transporter or pore. Glycerol is 1,2,3-propanetriol, a sweet, hygroscopic, viscous liquid, widely distributed in nature as a constituent of many lipids.
renal water transport
The directed movement of water (H2O) by the kidney.
renal system process involved in regulation of systemic arterial blood pressure
Renal process that modulates the force with which blood travels through the circulatory system. The process is controlled by a balance of processes that increase pressure and decrease pressure.
sphingomyelin metabolic process
The chemical reactions and pathways involving sphingomyelin, N-acyl-4-sphingenyl-1-O-phosphorylcholine, any of a class of phospholipids in which the amino group of sphingosine is in amide linkage with one of several fatty acids, while the terminal hydroxyl group of sphingosine is esterified to phosphorylcholine.
cellular response to mercury ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mercury ion stimulus.
cellular response to copper ion
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a copper ion stimulus.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
basolateral plasma membrane
The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
metal ion transmembrane transporter activity
Catalysis of the transfer of metal ions from one side of a membrane to the other.
phospholipase activity
Catalysis of the hydrolysis of a glycerophospholipid.
sphingomyelin phosphodiesterase activity
Catalysis of the reaction: sphingomyelin + H2O = N-acylsphingosine + choline phosphate.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
parathyroid hormone receptor activity
Combining with parathyroid hormone to initiate a change in cell activity.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
water transmembrane transporter activity
Enables the directed movement of water (H2O) from one side of a membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
inorganic anion exchanger activity
NA
phosphoric diester hydrolase activity
Catalysis of the hydrolysis of a phosphodiester to give a phosphomonoester and a free hydroxyl group.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
fucosyltransferase activity
Catalysis of the transfer of a fucosyl group to an acceptor molecule, typically another carbohydrate or a lipid.
anion transmembrane transporter activity
Catalysis of the transfer of a negatively charged ion from one side of a membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
polyol transmembrane transporter activity
Catalysis of the transfer of a polyol from one side of the membrane to the other. A polyol is any polyhydric alcohol.
glycerol transmembrane transporter activity
Catalysis of the transfer of glycerol from one side of the membrane to the other. Glycerol is 1,2,3-propanetriol, a sweet, hygroscopic, viscous liquid, widely distributed in nature as a constituent of many lipids.
water channel activity
Transport systems of this type catalyze facilitated diffusion of water (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
symporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported together in the same direction in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
antiporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported in opposite directions in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
solute:solute antiporter activity
Catalysis of the reaction: solute A(out) + solute B(in) = solute A(in) + solute B(out).
anion:anion antiporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion A(out) + anion B(in) = anion A(in) + anion B(out).
anion exchanger activity
NA
alcohol transmembrane transporter activity
Catalysis of the transfer of an alcohol from one side of the membrane to the other. An alcohol is any carbon compound that contains a hydroxyl group.
lipase activity
Catalysis of the hydrolysis of a lipid or phospholipid.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring glycosyl groups
Catalysis of the transfer of a glycosyl group from one compound (donor) to another (acceptor).
transferase activity, transferring hexosyl groups
Catalysis of the transfer of a hexosyl group from one compound (donor) to another (acceptor).
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on ester bonds
Catalysis of the hydrolysis of any ester bond.
3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity
Catalysis of the reaction: GDP-L-fucose + beta-D-galactosyl-(1,3)-N-acetyl-D-glucosaminyl-R = GDP + beta-D-galactosyl-(1,3)-[alpha-L-fucosyl-(1,4)]-N-acetyl-D-glucosaminyl-R.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
inorganic cation transmembrane transporter activity
Catalysis of the transfer of inorganic cations from one side of a membrane to the other. Inorganic cations are atoms or small molecules with a positive charge that do not contain carbon in covalent linkage.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
phosphoric ester hydrolase activity
Catalysis of the reaction: RPO-R' + H2O = RPOOH + R'H. This reaction is the hydrolysis of any phosphoric ester bond, any ester formed from orthophosphoric acid, O=P(OH)3.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
water channel activity
Transport systems of this type catalyze facilitated diffusion of water (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
sphingomyelin phosphodiesterase activity
Catalysis of the reaction: sphingomyelin + H2O = N-acylsphingosine + choline phosphate.
anion:anion antiporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion A(out) + anion B(in) = anion A(in) + anion B(out).
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00601 | 2.521e-02 | 0.2229 | 3 | 19 | Glycosphingolipid biosynthesis - lacto and neolacto series |
ACOT11acyl-CoA thioesterase 11 (ENSG00000162390), score: 0.49 ACPPacid phosphatase, prostate (ENSG00000014257), score: 0.58 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (ENSG00000006534), score: 0.58 ANO10anoctamin 10 (ENSG00000160746), score: 0.49 AP1M2adaptor-related protein complex 1, mu 2 subunit (ENSG00000129354), score: 0.5 APCDD1Ladenomatosis polyposis coli down-regulated 1-like (ENSG00000198768), score: 0.6 APEX2APEX nuclease (apurinic/apyrimidinic endonuclease) 2 (ENSG00000169188), score: 0.55 AQP2aquaporin 2 (collecting duct) (ENSG00000167580), score: 0.58 AQP3aquaporin 3 (Gill blood group) (ENSG00000165272), score: 0.5 ATP12AATPase, H+/K+ transporting, nongastric, alpha polypeptide (ENSG00000075673), score: 0.71 ATP6V1B1ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 (ENSG00000116039), score: 0.51 B3GNT3UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 (ENSG00000179913), score: 0.87 B4GALNT2beta-1,4-N-acetyl-galactosaminyl transferase 2 (ENSG00000167080), score: 0.57 C19orf21chromosome 19 open reading frame 21 (ENSG00000099812), score: 0.5 C1orf186chromosome 1 open reading frame 186 (ENSG00000196533), score: 1 C2orf24chromosome 2 open reading frame 24 (ENSG00000115649), score: 0.49 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.52 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.5 CALCAcalcitonin-related polypeptide alpha (ENSG00000110680), score: 0.54 CCNT2cyclin T2 (ENSG00000082258), score: -0.56 CDH16cadherin 16, KSP-cadherin (ENSG00000166589), score: 0.53 CDHR2cadherin-related family member 2 (ENSG00000074276), score: 0.56 CDHR5cadherin-related family member 5 (ENSG00000099834), score: 0.49 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (ENSG00000110172), score: -0.54 CHP2calcineurin B homologous protein 2 (ENSG00000166869), score: 0.65 CLCNKAchloride channel Ka (ENSG00000186510), score: 0.5 CLDN19claudin 19 (ENSG00000164007), score: 0.57 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.51 CRYAAcrystallin, alpha A (ENSG00000160202), score: 0.51 CRYBB3crystallin, beta B3 (ENSG00000100053), score: 0.56 CTRCchymotrypsin C (caldecrin) (ENSG00000162438), score: 0.56 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.5 CWH43cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) (ENSG00000109182), score: 0.55 CYP27B1cytochrome P450, family 27, subfamily B, polypeptide 1 (ENSG00000111012), score: 0.65 DNAJC11DnaJ (Hsp40) homolog, subfamily C, member 11 (ENSG00000007923), score: 0.53 EEF1Geukaryotic translation elongation factor 1 gamma (ENSG00000149016), score: 0.5 EIF3Keukaryotic translation initiation factor 3, subunit K (ENSG00000178982), score: 0.5 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.55 ERP27endoplasmic reticulum protein 27 (ENSG00000139055), score: 0.73 EVPLenvoplakin (ENSG00000167880), score: 0.49 FAM150Bfamily with sequence similarity 150, member B (ENSG00000189292), score: 0.51 FAM151Afamily with sequence similarity 151, member A (ENSG00000162391), score: 0.57 FCAMRFc receptor, IgA, IgM, high affinity (ENSG00000162897), score: 0.54 FOLR1folate receptor 1 (adult) (ENSG00000110195), score: 0.58 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (ENSG00000171124), score: 0.72 FUT6fucosyltransferase 6 (alpha (1,3) fucosyltransferase) (ENSG00000156413), score: 0.5 GGT6gamma-glutamyltransferase 6 (ENSG00000167741), score: 0.74 GNPDA1glucosamine-6-phosphate deaminase 1 (ENSG00000113552), score: 0.54 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 0.82 HEPACAM2HEPACAM family member 2 (ENSG00000188175), score: 0.5 HEXAhexosaminidase A (alpha polypeptide) (ENSG00000213614), score: 0.52 HHLA2HERV-H LTR-associating 2 (ENSG00000114455), score: 0.67 HOXC10homeobox C10 (ENSG00000180818), score: 0.53 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: 0.5 IL17RBinterleukin 17 receptor B (ENSG00000056736), score: 0.58 IL9Rinterleukin 9 receptor (ENSG00000124334), score: 0.56 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.51 INMTindolethylamine N-methyltransferase (ENSG00000106125), score: 0.63 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.5 KCNE3potassium voltage-gated channel, Isk-related family, member 3 (ENSG00000175538), score: 0.67 KCNH6potassium voltage-gated channel, subfamily H (eag-related), member 6 (ENSG00000173826), score: 0.82 KIAA0100KIAA0100 (ENSG00000007202), score: 0.53 KIAA1191KIAA1191 (ENSG00000122203), score: 0.52 LGALS2lectin, galactoside-binding, soluble, 2 (ENSG00000100079), score: 0.5 LYG1lysozyme G-like 1 (ENSG00000144214), score: 0.62 MCATmalonyl CoA:ACP acyltransferase (mitochondrial) (ENSG00000100294), score: 0.52 MCCD1mitochondrial coiled-coil domain 1 (ENSG00000204511), score: 0.69 METTL1methyltransferase like 1 (ENSG00000037897), score: 0.5 MIOXmyo-inositol oxygenase (ENSG00000100253), score: 0.49 MTNR1Amelatonin receptor 1A (ENSG00000168412), score: 0.5 MUC13mucin 13, cell surface associated (ENSG00000173702), score: 0.7 MUC20mucin 20, cell surface associated (ENSG00000176945), score: 0.52 NCCRP1non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) (ENSG00000188505), score: 0.76 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.55 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.5 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.67 PAX2paired box 2 (ENSG00000075891), score: 0.53 PAX8paired box 8 (ENSG00000125618), score: 0.49 PDZK1IP1PDZK1 interacting protein 1 (ENSG00000162366), score: 0.53 PEPDpeptidase D (ENSG00000124299), score: 0.53 PLA2G4Fphospholipase A2, group IVF (ENSG00000168907), score: 0.51 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.51 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (ENSG00000111725), score: 0.53 PRLRprolactin receptor (ENSG00000113494), score: 0.53 PRR13proline rich 13 (ENSG00000205352), score: 0.5 PRR15proline rich 15 (ENSG00000176532), score: 0.5 PRSS8protease, serine, 8 (ENSG00000052344), score: 0.52 PRXperiaxin (ENSG00000105227), score: 0.55 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.49 PTH1Rparathyroid hormone 1 receptor (ENSG00000160801), score: 0.53 PTH2Rparathyroid hormone 2 receptor (ENSG00000144407), score: 0.58 PTPN1protein tyrosine phosphatase, non-receptor type 1 (ENSG00000196396), score: 0.5 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.82 RDH8retinol dehydrogenase 8 (all-trans) (ENSG00000080511), score: 0.57 REG3Aregenerating islet-derived 3 alpha (ENSG00000172016), score: 0.54 RENrenin (ENSG00000143839), score: 0.57 RENBPrenin binding protein (ENSG00000102032), score: 0.5 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.49 S100A2S100 calcium binding protein A2 (ENSG00000196754), score: 0.6 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.54 SHISA3shisa homolog 3 (Xenopus laevis) (ENSG00000178343), score: 0.56 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (ENSG00000070915), score: 0.52 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (ENSG00000088386), score: 0.54 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.49 SLC22A11solute carrier family 22 (organic anion/urate transporter), member 11 (ENSG00000168065), score: 0.49 SLC22A12solute carrier family 22 (organic anion/urate transporter), member 12 (ENSG00000197891), score: 0.5 SLC22A13solute carrier family 22 (organic anion transporter), member 13 (ENSG00000172940), score: 0.7 SLC22A8solute carrier family 22 (organic anion transporter), member 8 (ENSG00000149452), score: 0.55 SLC30A2solute carrier family 30 (zinc transporter), member 2 (ENSG00000158014), score: 0.58 SLC34A3solute carrier family 34 (sodium phosphate), member 3 (ENSG00000198569), score: 0.49 SLC39A4solute carrier family 39 (zinc transporter), member 4 (ENSG00000147804), score: 0.57 SLC39A5solute carrier family 39 (metal ion transporter), member 5 (ENSG00000139540), score: 0.58 SLC44A4solute carrier family 44, member 4 (ENSG00000204385), score: 0.54 SLC4A1solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) (ENSG00000004939), score: 0.52 SLC5A10solute carrier family 5 (sodium/glucose cotransporter), member 10 (ENSG00000154025), score: 0.57 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.52 SLC5A2solute carrier family 5 (sodium/glucose cotransporter), member 2 (ENSG00000140675), score: 0.52 SLC5A8solute carrier family 5 (iodide transporter), member 8 (ENSG00000139357), score: 0.57 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.57 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (ENSG00000155465), score: 0.5 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.57 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (ENSG00000166311), score: 0.5 SMPDL3Bsphingomyelin phosphodiesterase, acid-like 3B (ENSG00000130768), score: 0.64 SOSTsclerostin (ENSG00000167941), score: 0.57 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.49 SUCLG1succinate-CoA ligase, alpha subunit (ENSG00000163541), score: 0.5 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.66 TACSTD2tumor-associated calcium signal transducer 2 (ENSG00000184292), score: 0.58 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.52 TM7SF3transmembrane 7 superfamily member 3 (ENSG00000064115), score: 0.5 TMC4transmembrane channel-like 4 (ENSG00000167608), score: 0.58 TMED4transmembrane emp24 protein transport domain containing 4 (ENSG00000158604), score: 0.61 TMEM150Btransmembrane protein 150B (ENSG00000180061), score: 0.67 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.52 TMEM174transmembrane protein 174 (ENSG00000164325), score: 0.51 TMEM72transmembrane protein 72 (ENSG00000187783), score: 0.53 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.63 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.63 TRIM10tripartite motif-containing 10 (ENSG00000204613), score: 0.8 TRIM15tripartite motif-containing 15 (ENSG00000204610), score: 0.82 TRIP6thyroid hormone receptor interactor 6 (ENSG00000087077), score: 0.5 TRPC7transient receptor potential cation channel, subfamily C, member 7 (ENSG00000069018), score: 0.86 TSPAN33tetraspanin 33 (ENSG00000158457), score: 0.52 TSPO2translocator protein 2 (ENSG00000112212), score: 0.58 TTC22tetratricopeptide repeat domain 22 (ENSG00000006555), score: 0.55 TUBAL3tubulin, alpha-like 3 (ENSG00000178462), score: 0.83 UNC5CLunc-5 homolog C (C. elegans)-like (ENSG00000124602), score: 0.51 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.66 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.49 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.55
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |