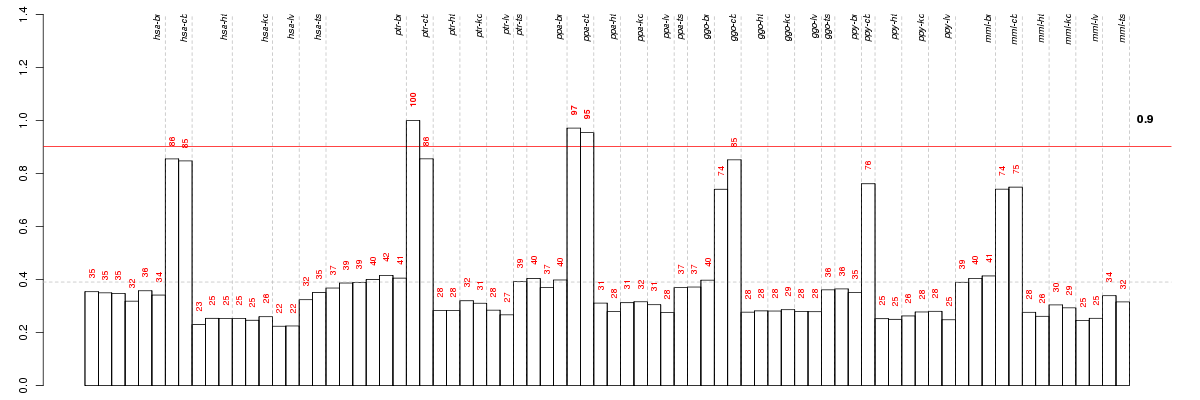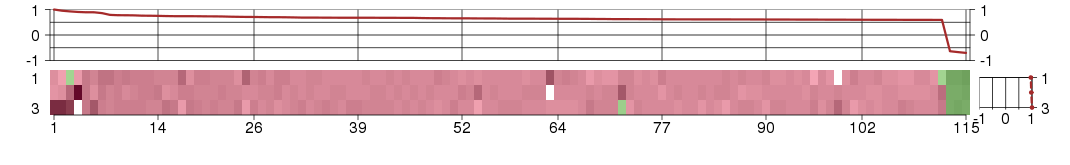

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.


ADAM10ADAM metallopeptidase domain 10 (ENSG00000137845), score: 0.6 AMIGO3adhesion molecule with Ig-like domain 3 (ENSG00000176020), score: 0.76 ARSJarylsulfatase family, member J (ENSG00000180801), score: 0.61 ATF7activating transcription factor 7 (ENSG00000170653), score: 0.71 BANK1B-cell scaffold protein with ankyrin repeats 1 (ENSG00000153064), score: 0.89 BARHL2BarH-like homeobox 2 (ENSG00000143032), score: 0.73 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.59 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.61 C8orf34chromosome 8 open reading frame 34 (ENSG00000165084), score: 0.74 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.68 CBLN3cerebellin 3 precursor (ENSG00000139899), score: 0.68 CCNJLcyclin J-like (ENSG00000135083), score: 0.6 CDH15cadherin 15, type 1, M-cadherin (myotubule) (ENSG00000129910), score: 0.67 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.68 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.61 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: 0.61 CER1cerberus 1, cysteine knot superfamily, homolog (Xenopus laevis) (ENSG00000147869), score: 0.64 CERKLceramide kinase-like (ENSG00000188452), score: 0.6 CHD7chromodomain helicase DNA binding protein 7 (ENSG00000171316), score: 0.65 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.6 CNTN6contactin 6 (ENSG00000134115), score: 0.61 COL13A1collagen, type XIII, alpha 1 (ENSG00000197467), score: 0.76 COL19A1collagen, type XIX, alpha 1 (ENSG00000082293), score: 0.64 CPLX4complexin 4 (ENSG00000166569), score: 0.93 CRLF3cytokine receptor-like factor 3 (ENSG00000176390), score: 0.77 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.73 DCLRE1BDNA cross-link repair 1B (ENSG00000118655), score: 0.64 DEFB136defensin, beta 136 (ENSG00000205884), score: 0.62 DGCR8DiGeorge syndrome critical region gene 8 (ENSG00000128191), score: 0.59 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.6 ECE1endothelin converting enzyme 1 (ENSG00000117298), score: -0.7 EN2engrailed homeobox 2 (ENSG00000164778), score: 0.71 EOMESeomesodermin (ENSG00000163508), score: 0.68 ESYT3extended synaptotagmin-like protein 3 (ENSG00000158220), score: 0.61 EXOSC10exosome component 10 (ENSG00000171824), score: 0.61 EXPH5exophilin 5 (ENSG00000110723), score: 0.68 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.69 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.61 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.86 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.63 FSTL5follistatin-like 5 (ENSG00000168843), score: 0.62 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.65 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.61 GCM2glial cells missing homolog 2 (Drosophila) (ENSG00000124827), score: 0.74 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.64 GPATCH8G patch domain containing 8 (ENSG00000186566), score: 0.6 GPRIN3GPRIN family member 3 (ENSG00000185477), score: 0.62 GRID2IPglutamate receptor, ionotropic, delta 2 (Grid2) interacting protein (ENSG00000215045), score: 0.74 GRM4glutamate receptor, metabotropic 4 (ENSG00000124493), score: 0.61 INO80INO80 homolog (S. cerevisiae) (ENSG00000128908), score: 0.61 IP6K2inositol hexakisphosphate kinase 2 (ENSG00000068745), score: 0.66 JMJD1Cjumonji domain containing 1C (ENSG00000171988), score: 0.68 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.7 KDM4Clysine (K)-specific demethylase 4C (ENSG00000107077), score: 0.64 KIAA0182KIAA0182 (ENSG00000131149), score: 0.64 KRT39keratin 39 (ENSG00000196859), score: 1 KRT40keratin 40 (ENSG00000204889), score: 0.95 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.62 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.74 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.67 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.68 MAMSTRMEF2 activating motif and SAP domain containing transcriptional regulator (ENSG00000176909), score: 0.6 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.67 MRGPRDMAS-related GPR, member D (ENSG00000172938), score: 0.71 MTBPMdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa (ENSG00000172167), score: 0.67 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.61 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (ENSG00000166886), score: 0.65 NHLH2nescient helix loop helix 2 (ENSG00000177551), score: 0.63 NHSL2NHS-like 2 (ENSG00000204131), score: 0.68 NKAIN1Na+/K+ transporting ATPase interacting 1 (ENSG00000084628), score: 0.64 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.79 NOTOnotochord homeobox (ENSG00000214513), score: 0.9 ODZ1odz, odd Oz/ten-m homolog 1(Drosophila) (ENSG00000009694), score: 0.65 PAX6paired box 6 (ENSG00000007372), score: 0.62 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.6 PISDphosphatidylserine decarboxylase (ENSG00000241878), score: 0.64 PKIBprotein kinase (cAMP-dependent, catalytic) inhibitor beta (ENSG00000135549), score: 0.72 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.63 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.61 PWWP2APWWP domain containing 2A (ENSG00000170234), score: 0.78 PYDC1PYD (pyrin domain) containing 1 (ENSG00000169900), score: 0.61 QSOX2quiescin Q6 sulfhydryl oxidase 2 (ENSG00000165661), score: 0.62 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.64 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (ENSG00000009413), score: 0.75 RFTN1raftlin, lipid raft linker 1 (ENSG00000131378), score: -0.64 RTEL1regulator of telomere elongation helicase 1 (ENSG00000026036), score: 0.6 SETD5SET domain containing 5 (ENSG00000168137), score: 0.66 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.68 SLC6A5solute carrier family 6 (neurotransmitter transporter, glycine), member 5 (ENSG00000165970), score: 0.59 SNRKSNF related kinase (ENSG00000163788), score: 0.7 SP4Sp4 transcription factor (ENSG00000105866), score: 0.59 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.6 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.69 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.74 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.61 TGM1transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) (ENSG00000092295), score: 0.64 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.63 TLL1tolloid-like 1 (ENSG00000038295), score: 0.73 TMC2transmembrane channel-like 2 (ENSG00000149488), score: 0.89 TP73tumor protein p73 (ENSG00000078900), score: 0.59 TRANK1tetratricopeptide repeat and ankyrin repeat containing 1 (ENSG00000168016), score: 0.6 TRIM11tripartite motif-containing 11 (ENSG00000154370), score: 0.77 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.68 UNC13Bunc-13 homolog B (C. elegans) (ENSG00000198722), score: -0.67 UPF0639UPF0639 protein (ENSG00000175985), score: 0.59 USTuronyl-2-sulfotransferase (ENSG00000111962), score: 0.65 VSX1visual system homeobox 1 (ENSG00000100987), score: 0.7 XKR7XK, Kell blood group complex subunit-related family, member 7 (ENSG00000101321), score: 0.62 ZIC2Zic family member 2 (odd-paired homolog, Drosophila) (ENSG00000043355), score: 0.6 ZIC4Zic family member 4 (ENSG00000174963), score: 0.68 ZIC5Zic family member 5 (odd-paired homolog, Drosophila) (ENSG00000139800), score: 0.66 ZNF337zinc finger protein 337 (ENSG00000130684), score: 0.66 ZNF397zinc finger protein 397 (ENSG00000186812), score: 0.6 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.61
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |