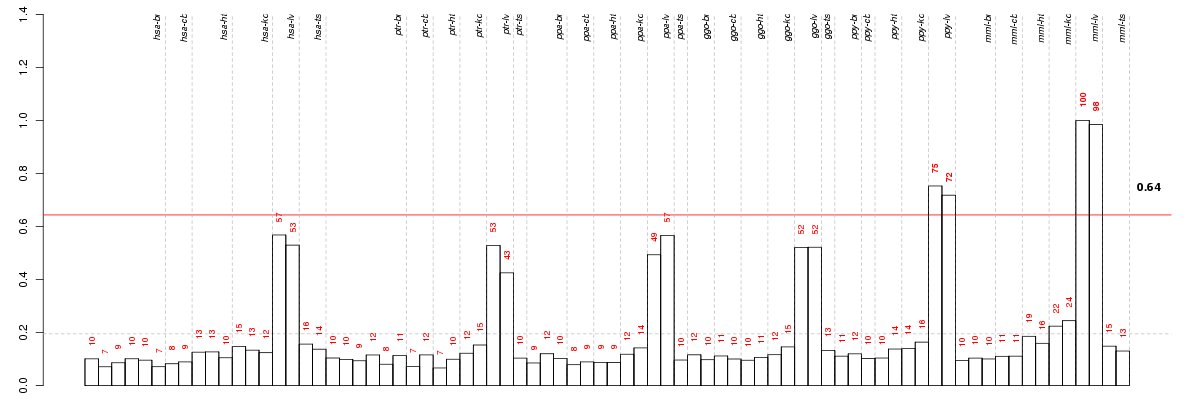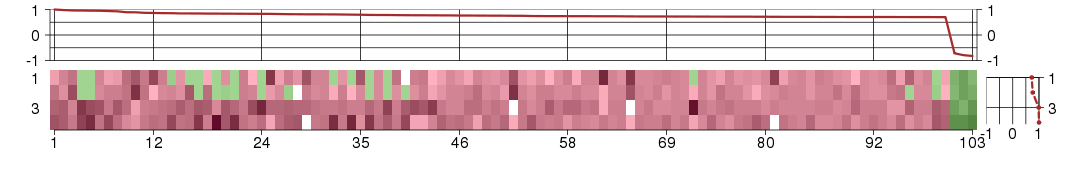

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
immune effector process
Any process of the immune system that occurs as part of an immune response.
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies). An example of this is the adaptive immune response found in Mus musculus.
acute inflammatory response
Inflammation which comprises a rapid, short-lived, relatively uniform response to acute injury or antigenic challenge and is characterized by accumulations of fluid, plasma proteins, and granulocytic leukocytes. An acute inflammatory response occurs within a matter of minutes or hours, and either resolves within a few days or becomes a chronic inflammatory response.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
protein maturation by peptide bond cleavage
The hydrolysis of a peptide bond or bonds within a protein as part of protein maturation, the process leading to the attainment of the full functional capacity of a protein.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
humoral immune response
An immune response mediated through a body fluid.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
immunoglobulin mediated immune response
An immune response mediated by immunoglobulins, whether cell-bound or in solution.
protein processing
Any protein maturation process achieved by the cleavage of peptide bonds.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
NA
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
complement activation, alternative pathway
Any process involved in the activation of any of the steps of the alternative pathway of the complement cascade which allows for the direct killing of microbes and the regulation of other immune processes.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
complement activation, classical pathway
Any process involved in the activation of any of the steps of the classical pathway of the complement cascade which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cell surface binding
Interacting selectively and non-covalently with any component on the surface of a cell.
bacterial cell surface binding
Interacting selectively and non-covalently with any component on the surface of a bacterial cell.
Gram-negative bacterial cell surface binding
Interacting selectively and non-covalently with any component on the surface of a Gram-negative bacterial cell.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00140 | 2.411e-02 | 0.2179 | 3 | 22 | Steroid hormone biosynthesis |
| 04610 | 2.647e-02 | 0.4953 | 4 | 50 | Complement and coagulation cascades |
| 05020 | 2.939e-02 | 0.2377 | 3 | 24 | Prion diseases |
| 00830 | 3.211e-02 | 0.2477 | 3 | 25 | Retinol metabolism |
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.86 ADAM12ADAM metallopeptidase domain 12 (ENSG00000148848), score: 0.84 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.87 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.74 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.76 AOAHacyloxyacyl hydrolase (neutrophil) (ENSG00000136250), score: 0.72 APOA4apolipoprotein A-IV (ENSG00000110244), score: 0.81 APOFapolipoprotein F (ENSG00000175336), score: 0.83 AQP8aquaporin 8 (ENSG00000103375), score: 0.96 ARL2BPADP-ribosylation factor-like 2 binding protein (ENSG00000102931), score: -0.79 BPIL1bactericidal/permeability-increasing protein-like 1 (ENSG00000078898), score: 0.89 C17orf87chromosome 17 open reading frame 87 (ENSG00000161929), score: 0.93 C2CD4AC2 calcium-dependent domain containing 4A (ENSG00000198535), score: 0.7 C8Acomplement component 8, alpha polypeptide (ENSG00000157131), score: 0.73 C8Bcomplement component 8, beta polypeptide (ENSG00000021852), score: 0.71 C9complement component 9 (ENSG00000113600), score: 0.74 C9orf152chromosome 9 open reading frame 152 (ENSG00000188959), score: 0.85 CAMPcathelicidin antimicrobial peptide (ENSG00000164047), score: 0.79 CCL23chemokine (C-C motif) ligand 23 (ENSG00000167236), score: 0.76 CCL24chemokine (C-C motif) ligand 24 (ENSG00000106178), score: 0.84 CD1CCD1c molecule (ENSG00000158481), score: 0.71 CD209CD209 molecule (ENSG00000090659), score: 0.89 CD33CD33 molecule (ENSG00000105383), score: 0.85 CD5LCD5 molecule-like (ENSG00000073754), score: 0.76 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.82 CFPcomplement factor properdin (ENSG00000126759), score: 0.76 CLDN14claudin 14 (ENSG00000159261), score: 0.77 CPA6carboxypeptidase A6 (ENSG00000165078), score: 0.72 CRISP3cysteine-rich secretory protein 3 (ENSG00000096006), score: 0.83 CRYBA2crystallin, beta A2 (ENSG00000163499), score: 0.7 CXCR2chemokine (C-X-C motif) receptor 2 (ENSG00000180871), score: 0.95 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.74 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (ENSG00000021461), score: 0.72 DNASE2Bdeoxyribonuclease II beta (ENSG00000137976), score: 0.74 DSC3desmocollin 3 (ENSG00000134762), score: 0.83 EMR1egf-like module containing, mucin-like, hormone receptor-like 1 (ENSG00000174837), score: 0.83 ENDOGendonuclease G (ENSG00000167136), score: 0.71 FETUBfetuin B (ENSG00000090512), score: 0.81 FGD2FYVE, RhoGEF and PH domain containing 2 (ENSG00000146192), score: 0.82 FOXA1forkhead box A1 (ENSG00000129514), score: 0.76 FOXP3forkhead box P3 (ENSG00000049768), score: 1 FSTfollistatin (ENSG00000134363), score: 0.74 GDF2growth differentiation factor 2 (ENSG00000128802), score: 0.7 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.81 GPRC5DG protein-coupled receptor, family C, group 5, member D (ENSG00000111291), score: 0.74 HRASLS2HRAS-like suppressor 2 (ENSG00000133328), score: 0.7 IGFALSinsulin-like growth factor binding protein, acid labile subunit (ENSG00000099769), score: 0.71 IL1R2interleukin 1 receptor, type II (ENSG00000115590), score: 0.81 KRT27keratin 27 (ENSG00000171446), score: 0.75 KRT28keratin 28 (ENSG00000173908), score: 0.96 LBPlipopolysaccharide binding protein (ENSG00000129988), score: 0.71 LIPMlipase, family member M (ENSG00000173239), score: 0.72 LONP2lon peptidase 2, peroxisomal (ENSG00000102910), score: 0.71 LRRC25leucine rich repeat containing 25 (ENSG00000175489), score: 0.71 LRRC8Eleucine rich repeat containing 8 family, member E (ENSG00000171017), score: 0.71 MASP2mannan-binding lectin serine peptidase 2 (ENSG00000009724), score: 0.72 MLANAmelan-A (ENSG00000120215), score: 0.73 MOCOSmolybdenum cofactor sulfurase (ENSG00000075643), score: 0.72 NLRP12NLR family, pyrin domain containing 12 (ENSG00000142405), score: 0.71 ONECUT1one cut homeobox 1 (ENSG00000169856), score: 0.8 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.78 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: 0.72 PFKFB16-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (ENSG00000158571), score: 0.75 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.77 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.86 PRHOXNBparahox cluster neighbor (ENSG00000183463), score: 0.95 PROZprotein Z, vitamin K-dependent plasma glycoprotein (ENSG00000126231), score: 0.71 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.82 PSKH2protein serine kinase H2 (ENSG00000147613), score: 0.78 PTCRApre T-cell antigen receptor alpha (ENSG00000171611), score: 0.86 RDH16retinol dehydrogenase 16 (all-trans) (ENSG00000139547), score: 0.71 RGS13regulator of G-protein signaling 13 (ENSG00000127074), score: 0.84 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.77 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.72 SERPINA11serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 (ENSG00000186910), score: 0.7 SERPINA12serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (ENSG00000165953), score: 0.8 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (ENSG00000123561), score: 0.74 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (ENSG00000088827), score: 0.72 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (ENSG00000100652), score: 0.73 SLCO1B1solute carrier organic anion transporter family, member 1B1 (ENSG00000134538), score: 0.73 SNX20sorting nexin 20 (ENSG00000167208), score: 0.76 SOAT2sterol O-acyltransferase 2 (ENSG00000167780), score: 0.94 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.73 SRPRsignal recognition particle receptor (docking protein) (ENSG00000182934), score: 0.71 STAB2stabilin 2 (ENSG00000136011), score: 0.74 SYVN1synovial apoptosis inhibitor 1, synoviolin (ENSG00000162298), score: 0.73 TATtyrosine aminotransferase (ENSG00000198650), score: 0.72 TECTBtectorin beta (ENSG00000119913), score: 0.85 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (ENSG00000104953), score: 0.7 TM7SF2transmembrane 7 superfamily member 2 (ENSG00000149809), score: 0.72 TMEM154transmembrane protein 154 (ENSG00000170006), score: 0.77 TNFAIP8L2tumor necrosis factor, alpha-induced protein 8-like 2 (ENSG00000163154), score: 0.74 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.81 TRPM5transient receptor potential cation channel, subfamily M, member 5 (ENSG00000070985), score: 0.84 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.73 TTPALtocopherol (alpha) transfer protein-like (ENSG00000124120), score: 0.71 UROC1urocanase domain containing 1 (ENSG00000159650), score: 0.71 VMO1vitelline membrane outer layer 1 homolog (chicken) (ENSG00000182853), score: 0.78 VWCEvon Willebrand factor C and EGF domains (ENSG00000167992), score: 0.77 XDHxanthine dehydrogenase (ENSG00000158125), score: 0.74
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppy_lv_f_ca1 | ppy | lv | f | _ |
| ppy_lv_m_ca1 | ppy | lv | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |