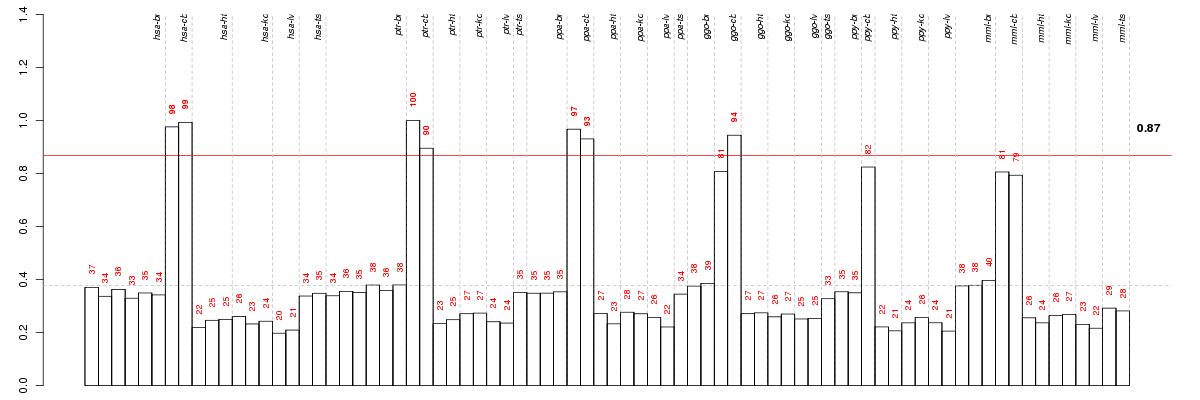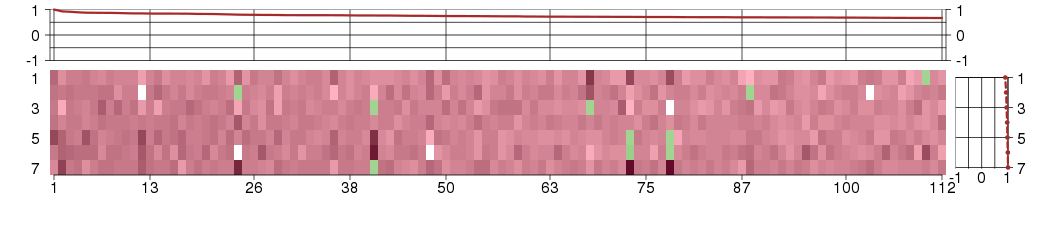

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
brain development
The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
neurogenesis
Generation of cells within the nervous system.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
midbrain development
The process whose specific outcome is the progression of the midbrain over time, from its formation to the mature structure. The midbrain is the middle division of the three primary divisions of the developing chordate brain or the corresponding part of the adult brain (in vertebrates, includes a ventral part containing the cerebral peduncles and a dorsal tectum containing the corpora quadrigemina and that surrounds the aqueduct of Sylvius connecting the third and fourth ventricles).
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
neurogenesis
Generation of cells within the nervous system.
brain development
The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.).
midbrain development
The process whose specific outcome is the progression of the midbrain over time, from its formation to the mature structure. The midbrain is the middle division of the three primary divisions of the developing chordate brain or the corresponding part of the adult brain (in vertebrates, includes a ventral part containing the cerebral peduncles and a dorsal tectum containing the corpora quadrigemina and that surrounds the aqueduct of Sylvius connecting the third and fourth ventricles).
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.

cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
all
NA

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
sequence-specific DNA binding transcription factor activity
Interacting selectively and non-covalently with a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
sequence-specific DNA binding
Interacting selectively and non-covalently with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding.
all
NA
ANKLE1ankyrin repeat and LEM domain containing 1 (ENSG00000160117), score: 0.79 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.71 BANK1B-cell scaffold protein with ankyrin repeats 1 (ENSG00000153064), score: 0.75 BARHL1BarH-like homeobox 1 (ENSG00000125492), score: 0.84 BARHL2BarH-like homeobox 2 (ENSG00000143032), score: 0.89 BTN2A1butyrophilin, subfamily 2, member A1 (ENSG00000112763), score: 0.7 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.71 C16orf11chromosome 16 open reading frame 11 (ENSG00000161992), score: 0.76 C20orf117chromosome 20 open reading frame 117 (ENSG00000149639), score: 0.69 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.68 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.78 CBLN3cerebellin 3 precursor (ENSG00000139899), score: 0.78 CCNJLcyclin J-like (ENSG00000135083), score: 0.68 CDH15cadherin 15, type 1, M-cadherin (myotubule) (ENSG00000129910), score: 0.87 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.71 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.72 CECR2cat eye syndrome chromosome region, candidate 2 (ENSG00000099954), score: 0.68 CERKLceramide kinase-like (ENSG00000188452), score: 0.69 CHD7chromodomain helicase DNA binding protein 7 (ENSG00000171316), score: 0.77 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.78 CLK4CDC-like kinase 4 (ENSG00000113240), score: 0.69 CNPY1canopy 1 homolog (zebrafish) (ENSG00000146910), score: 0.71 COL13A1collagen, type XIII, alpha 1 (ENSG00000197467), score: 0.87 CPLX4complexin 4 (ENSG00000166569), score: 0.8 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.91 DCLRE1BDNA cross-link repair 1B (ENSG00000118655), score: 0.73 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.68 DUOX1dual oxidase 1 (ENSG00000137857), score: 0.73 EN2engrailed homeobox 2 (ENSG00000164778), score: 0.84 EOMESeomesodermin (ENSG00000163508), score: 0.75 ESYT3extended synaptotagmin-like protein 3 (ENSG00000158220), score: 0.69 EXPH5exophilin 5 (ENSG00000110723), score: 0.78 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.85 FGF17fibroblast growth factor 17 (ENSG00000158815), score: 0.69 FGF3fibroblast growth factor 3 (ENSG00000186895), score: 0.77 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.79 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.74 FSTL5follistatin-like 5 (ENSG00000168843), score: 0.71 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.72 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.67 GPRIN3GPRIN family member 3 (ENSG00000185477), score: 0.77 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.7 GRID2IPglutamate receptor, ionotropic, delta 2 (Grid2) interacting protein (ENSG00000215045), score: 0.84 GRIN2Cglutamate receptor, ionotropic, N-methyl D-aspartate 2C (ENSG00000161509), score: 0.7 GRM4glutamate receptor, metabotropic 4 (ENSG00000124493), score: 0.78 HINFPhistone H4 transcription factor (ENSG00000172273), score: 0.69 IFFO2intermediate filament family orphan 2 (ENSG00000169991), score: 0.72 IL16interleukin 16 (lymphocyte chemoattractant factor) (ENSG00000172349), score: 0.74 INCENPinner centromere protein antigens 135/155kDa (ENSG00000149503), score: 0.69 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.69 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.7 KDM4Clysine (K)-specific demethylase 4C (ENSG00000107077), score: 0.76 KIAA0182KIAA0182 (ENSG00000131149), score: 0.68 KIAA0802KIAA0802 (ENSG00000168502), score: 0.74 KRT39keratin 39 (ENSG00000196859), score: 0.71 KRT40keratin 40 (ENSG00000204889), score: 0.71 LBX1ladybird homeobox 1 (ENSG00000138136), score: 0.77 LHX5LIM homeobox 5 (ENSG00000089116), score: 0.74 LOC146429putative solute carrier family 22 member ENSG00000182157 (ENSG00000182157), score: 0.88 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.67 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.87 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.72 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.76 MAXMYC associated factor X (ENSG00000125952), score: 0.67 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.81 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.67 MRGPRDMAS-related GPR, member D (ENSG00000172938), score: 0.72 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.7 NHLH2nescient helix loop helix 2 (ENSG00000177551), score: 0.86 NKAIN1Na+/K+ transporting ATPase interacting 1 (ENSG00000084628), score: 0.72 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.84 NRIP2nuclear receptor interacting protein 2 (ENSG00000053702), score: 0.75 NYNRINNYN domain and retroviral integrase containing (ENSG00000205978), score: 0.67 ODZ1odz, odd Oz/ten-m homolog 1(Drosophila) (ENSG00000009694), score: 0.79 OTX2orthodenticle homeobox 2 (ENSG00000165588), score: 0.69 PAX6paired box 6 (ENSG00000007372), score: 0.72 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.7 PISDphosphatidylserine decarboxylase (ENSG00000241878), score: 0.73 PKIBprotein kinase (cAMP-dependent, catalytic) inhibitor beta (ENSG00000135549), score: 0.84 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.74 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.75 PWWP2APWWP domain containing 2A (ENSG00000170234), score: 0.71 PYDC1PYD (pyrin domain) containing 1 (ENSG00000169900), score: 0.79 QSOX2quiescin Q6 sulfhydryl oxidase 2 (ENSG00000165661), score: 0.69 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.83 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (ENSG00000009413), score: 0.67 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: 0.68 SCGNsecretagogin, EF-hand calcium binding protein (ENSG00000079689), score: 0.69 SETD5SET domain containing 5 (ENSG00000168137), score: 0.7 SGK223homolog of rat pragma of Rnd2 (ENSG00000182319), score: 0.67 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.83 SNRKSNF related kinase (ENSG00000163788), score: 0.72 SP4Sp4 transcription factor (ENSG00000105866), score: 0.69 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.85 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.69 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.71 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.72 TGM1transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) (ENSG00000092295), score: 0.77 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.74 TLL1tolloid-like 1 (ENSG00000038295), score: 0.82 TMC2transmembrane channel-like 2 (ENSG00000149488), score: 0.93 TP73tumor protein p73 (ENSG00000078900), score: 0.81 TRIM11tripartite motif-containing 11 (ENSG00000154370), score: 0.76 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.83 UBASH3Bubiquitin associated and SH3 domain containing B (ENSG00000154127), score: 0.68 VSX1visual system homeobox 1 (ENSG00000100987), score: 1 XKR7XK, Kell blood group complex subunit-related family, member 7 (ENSG00000101321), score: 0.78 ZIC2Zic family member 2 (odd-paired homolog, Drosophila) (ENSG00000043355), score: 0.74 ZIC4Zic family member 4 (ENSG00000174963), score: 0.85 ZIC5Zic family member 5 (odd-paired homolog, Drosophila) (ENSG00000139800), score: 0.78 ZNF296zinc finger protein 296 (ENSG00000170684), score: 0.71 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.77
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |