
Under-expression is coded with green,
over-expression with red color.

skeletal system development
The process whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure. The skeleton is the bony framework of the body in vertebrates (endoskeleton) or the hard outer envelope of insects (exoskeleton or dermoskeleton).
regionalization
The pattern specification process by which an axis or axes is subdivided in space to define an area or volume in which specific patterns of cell differentiation will take place or in which cells interpret a specific environment.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cation transport
The directed movement of cations, atoms or small molecules with a net positive charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
anion transport
The directed movement of anions, atoms or small molecules with a net negative charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
metal ion transport
The directed movement of metal ions, any metal ion with an electric charge, into, out of, within or between cells by means of some external agent such as a transporter or pore.
amino acid transport
The directed movement of amino acids, organic acids containing one or more amino substituents, into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
embryo development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
pattern specification process
Any developmental process that results in the creation of defined areas or spaces within an organism to which cells respond and eventually are instructed to differentiate.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
embryo development ending in birth or egg hatching
The process whose specific outcome is the progression of an embryo over time, from zygote formation until the end of the embryonic life stage. The end of the embryonic life stage is organism-specific and may be somewhat arbitrary; for mammals it is usually considered to be birth, for insects the hatching of the first instar larva from the eggshell.
anterior/posterior pattern formation
The regionalization process by which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism.
proximal/distal pattern formation
The regionalization process by which specific areas of cell differentiation are determined along a proximal/distal axis. The proximal/distal axis is defined by a line that runs from main body (proximal end) of an organism outward (distal end).
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to extracellular stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
monovalent inorganic cation transport
The directed movement of inorganic cations with a valency of one into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic cations are atoms or small molecules with a positive charge which do not contain carbon in covalent linkage.
inorganic anion transport
The directed movement of inorganic anions into, out of, within or between cells by means of some external agent such as a transporter or pore. Inorganic anions are atoms or small molecules with a negative charge which do not contain carbon in covalent linkage.
organic anion transport
The directed movement of organic anions into, out of, within or between cells by means of some external agent such as a transporter or pore. Organic anions are atoms or small molecules with a negative charge which contain carbon in covalent linkage.
amine transport
The directed movement of amines, including polyamines, organic compounds containing one or more amino groups, into, out of, within or between cells by means of some external agent such as a transporter or pore.
organic acid transport
The directed movement of organic acids, any acidic compound containing carbon in covalent linkage, into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
calcium-independent cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules that do not require the presence of calcium for the interaction.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
intracellular receptor mediated signaling pathway
Any series of molecular signals initiated by a ligand binding to an receptor located within a cell.
thyroid gland development
The process whose specific outcome is the progression of the thyroid gland over time, from its formation to the mature structure. The thyroid gland is an endoderm-derived gland that produces thyroid hormone.
mammary gland development
The process whose specific outcome is the progression of the mammary gland over time, from its formation to the mature structure. The mammary gland is a large compound sebaceous gland that in female mammals is modified to secrete milk. Its development starts with the formation of the mammary line and ends as the mature gland cycles between nursing and weaning stages.
response to nutrient levels
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
cellular response to nutrient levels
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
cellular response to nutrient
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
response to vitamin
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin stimulus.
response to vitamin D
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin D stimulus.
endocrine system development
Progression of the endocrine system over time, from its formation to a mature structure. The endocrine system is a system of hormones and ductless glands, where the glands release hormones directly into the blood, lymph or other intercellular fluid, and the hormones circulate within the body to affect distant organs. The major glands that make up the human endocrine system are the hypothalamus, pituitary, thyroid, parathryoids, adrenals, pineal body, and the reproductive glands which include the ovaries and testes.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
homeostatic process
Any biological process involved in the maintenance of an internal steady-state.
chordate embryonic development
The process whose specific outcome is the progression of the embryo over time, from zygote formation through a stage including a notochord and neural tube until birth or egg hatching.
carboxylic acid transport
The directed movement of carboxylic acids into, out of, within or between cells by means of some external agent such as a transporter or pore. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
embryonic skeletal system morphogenesis
The process by which the anatomical structures of the skeleton are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form.
skeletal system morphogenesis
The process by which the anatomical structures of the skeleton are generated and organized. Morphogenesis pertains to the creation of form.
embryonic skeletal system development
The process, occurring during the embryonic phase, whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
gland development
The process whose specific outcome is the progression of a gland over time, from its formation to the mature structure. A gland is an organ specialised for secretion.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
vitamin D receptor signaling pathway
The series of molecular signals generated as a consequence of a vitamin D receptor binding to one of its physiological ligands.
cellular response to chemical stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
cellular response to vitamin
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin stimulus.
cellular response to vitamin D
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin D stimulus.
cellular response to external stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
all
NA
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
embryo development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
pattern specification process
Any developmental process that results in the creation of defined areas or spaces within an organism to which cells respond and eventually are instructed to differentiate.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cellular response to chemical stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
transmembrane transport
The process whereby a solute is transported from one side of a membrane to the other. This process includes the actual movement of the solute, and any regulation and preparatory steps, such as reduction of the solute.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
cellular response to extracellular stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an extracellular stimulus.
cellular response to nutrient
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
cellular response to nutrient
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
skeletal system morphogenesis
The process by which the anatomical structures of the skeleton are generated and organized. Morphogenesis pertains to the creation of form.
embryonic skeletal system morphogenesis
The process by which the anatomical structures of the skeleton are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form.
embryonic skeletal system development
The process, occurring during the embryonic phase, whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure.
thyroid gland development
The process whose specific outcome is the progression of the thyroid gland over time, from its formation to the mature structure. The thyroid gland is an endoderm-derived gland that produces thyroid hormone.
response to nutrient
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nutrient stimulus.
cellular response to nutrient levels
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus reflecting the presence, absence, or concentration of nutrients.
cellular response to vitamin
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin stimulus.
amino acid transport
The directed movement of amino acids, organic acids containing one or more amino substituents, into, out of, within or between cells by means of some external agent such as a transporter or pore.
embryonic skeletal system morphogenesis
The process by which the anatomical structures of the skeleton are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form.
cellular response to vitamin D
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vitamin D stimulus.
sodium ion transport
The directed movement of sodium ions (Na+) into, out of, within or between cells by means of some external agent such as a transporter or pore.
vitamin D receptor signaling pathway
The series of molecular signals generated as a consequence of a vitamin D receptor binding to one of its physiological ligands.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
proton-transporting two-sector ATPase complex
A large protein complex that catalyzes the synthesis or hydrolysis of ATP by a rotational mechanism, coupled to the transport of protons across a membrane. The complex comprises a membrane sector (F0, V0, or A0) that carries out proton transport and a cytoplasmic compartment sector (F1, V1, or A1) that catalyzes ATP synthesis or hydrolysis. Two major types have been characterized: V-type ATPases couple ATP hydrolysis to the transport of protons across a concentration gradient, whereas F-type ATPases, also known as ATP synthases, normally run in the reverse direction to utilize energy from a proton concentration or electrochemical gradient to synthesize ATP. A third type, A-type ATPases have been found in archaea, and are closely related to eukaryotic V-type ATPases but are reversible.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell fraction
A generic term for parts of cells prepared by disruptive biochemical techniques.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane fraction
That fraction of cells, prepared by disruptive biochemical methods, that includes the plasma and other membranes.
insoluble fraction
That fraction of cells, prepared by disruptive biochemical methods, that is not soluble in water.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuolar membrane
The lipid bilayer surrounding the vacuole and separating its contents from the cytoplasm of the cell.
brush border
Dense covering of microvilli on the apical surface of epithelial cells in tissues such as the intestine, kidney, and choroid plexus; the microvilli aid absorption by increasing the surface area of the cell.
basolateral plasma membrane
The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
vacuolar proton-transporting V-type ATPase complex
A proton-transporting two-sector ATPase complex found in the vacuolar membrane, where it acts as a proton pump to mediate acidification of the vacuolar lumen.
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
proton-transporting V-type ATPase complex
A proton-transporting two-sector ATPase complex that couples ATP hydrolysis to the transport of protons across a concentration gradient. The resulting transmembrane electrochemical potential of H+ is used to drive a variety of (i) secondary active transport systems via H+-dependent symporters and antiporters and (ii) channel-mediated transport systems. The complex comprises a membrane sector (V0) that carries out proton transport and a cytoplasmic compartment sector (V1) that catalyzes ATP hydrolysis. V-type ATPases are found in the membranes of organelles such as vacuoles, endosomes, and lysosomes, and in the plasma membrane.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
proton-transporting two-sector ATPase complex
A large protein complex that catalyzes the synthesis or hydrolysis of ATP by a rotational mechanism, coupled to the transport of protons across a membrane. The complex comprises a membrane sector (F0, V0, or A0) that carries out proton transport and a cytoplasmic compartment sector (F1, V1, or A1) that catalyzes ATP synthesis or hydrolysis. Two major types have been characterized: V-type ATPases couple ATP hydrolysis to the transport of protons across a concentration gradient, whereas F-type ATPases, also known as ATP synthases, normally run in the reverse direction to utilize energy from a proton concentration or electrochemical gradient to synthesize ATP. A third type, A-type ATPases have been found in archaea, and are closely related to eukaryotic V-type ATPases but are reversible.
vacuolar membrane
The lipid bilayer surrounding the vacuole and separating its contents from the cytoplasm of the cell.
vacuolar proton-transporting V-type ATPase complex
A proton-transporting two-sector ATPase complex found in the vacuolar membrane, where it acts as a proton pump to mediate acidification of the vacuolar lumen.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
vacuole
A closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material. Cells contain one or several vacuoles, that may have different functions from each other. Vacuoles have a diverse array of functions. They can act as a storage organelle for nutrients or waste products, as a degradative compartment, as a cost-effective way of increasing cell size, and as a homeostatic regulator controlling both turgor pressure and pH of the cytosol.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cell projection membrane
The portion of the plasma membrane surrounding a cell surface projection.
brush border membrane
The portion of the plasma membrane surrounding the brush border.
vacuolar proton-transporting V-type ATPase complex
A proton-transporting two-sector ATPase complex found in the vacuolar membrane, where it acts as a proton pump to mediate acidification of the vacuolar lumen.
vacuolar part
Any constituent part of a vacuole, a closed structure, found only in eukaryotic cells, that is completely surrounded by unit membrane and contains liquid material.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
sequence-specific DNA binding transcription factor activity
Interacting selectively and non-covalently with a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
metal ion transmembrane transporter activity
Catalysis of the transfer of metal ions from one side of a membrane to the other.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
sodium channel activity
Catalysis of facilitated diffusion of a sodium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
amine transmembrane transporter activity
Catalysis of the transfer of amines, including polyamines, from one side of the membrane to the other. Amines are organic compounds that are weakly basic in character and contain an amino (-NH2) or substituted amino group.
amino acid transmembrane transporter activity
Catalysis of the transfer of amino acids from one side of a membrane to the other. Amino acids are organic molecules that contain an amino group and a carboxyl group.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
organic acid transmembrane transporter activity
Catalysis of the transfer of organic acids, any acidic compound containing carbon in covalent linkage, from one side of the membrane to the other.
secondary active transmembrane transporter activity
Catalysis of the transfer of a solute from one side of a membrane to the other, up its concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction and is driven by a chemiosmotic source of energy. Chemiosmotic sources of energy include uniport, symport or antiport.
inorganic anion exchanger activity
NA
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
anion transmembrane transporter activity
Catalysis of the transfer of a negatively charged ion from one side of a membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
monovalent inorganic cation transmembrane transporter activity
Catalysis of the transfer of a inorganic cations with a valency of one from one side of a membrane to the other. Inorganic cations are atoms or small molecules with a positive charge that do not contain carbon in covalent linkage.
sodium ion transmembrane transporter activity
Catalysis of the transfer of sodium ions (Na+) from one side of a membrane to the other.
organic cation transmembrane transporter activity
Catalysis of the transfer of organic cations from one side of a membrane to the other. Organic cations are atoms or small molecules with a positive charge that contain carbon in covalent linkage.
inorganic anion transmembrane transporter activity
Catalysis of the transfer of inorganic anions from one side of a membrane to the other. Inorganic anions are atoms or small molecules with a negative charge which do not contain carbon in covalent linkage.
phosphate transmembrane transporter activity
Catalysis of the transfer of phosphate (PO4 3-) ions from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
amiloride-sensitive sodium channel activity
NA
symporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported together in the same direction in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
solute:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: solute(out) + cation(out) = solute(in) + cation(in).
anion:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion(out) + cation(out) = anion(in) + cation(in).
antiporter activity
Enables the active transport of a solute across a membrane by a mechanism whereby two or more species are transported in opposite directions in a tightly coupled process not directly linked to a form of energy other than chemiosmotic energy.
solute:solute antiporter activity
Catalysis of the reaction: solute A(out) + solute B(in) = solute A(in) + solute B(out).
anion:anion antiporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion A(out) + anion B(in) = anion A(in) + anion B(out).
sodium-dependent phosphate transmembrane transporter activity
Catalysis of the transfer of phosphate (PO4 3-) ions from one side of a membrane to the other, requiring sodium ions.
anion exchanger activity
NA
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
inorganic cation transmembrane transporter activity
Catalysis of the transfer of inorganic cations from one side of a membrane to the other. Inorganic cations are atoms or small molecules with a positive charge that do not contain carbon in covalent linkage.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
sequence-specific DNA binding
Interacting selectively and non-covalently with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding.
carboxylic acid transmembrane transporter activity
Catalysis of the transfer of carboxylic acids from one side of the membrane to the other. Carboxylic acids are organic acids containing one or more carboxyl (COOH) groups or anions (COO-).
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
amine transmembrane transporter activity
Catalysis of the transfer of amines, including polyamines, from one side of the membrane to the other. Amines are organic compounds that are weakly basic in character and contain an amino (-NH2) or substituted amino group.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
amino acid transmembrane transporter activity
Catalysis of the transfer of amino acids from one side of a membrane to the other. Amino acids are organic molecules that contain an amino group and a carboxyl group.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
solute:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: solute(out) + cation(out) = solute(in) + cation(in).
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
anion:cation symporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion(out) + cation(out) = anion(in) + cation(in).
anion:anion antiporter activity
Catalysis of the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: anion A(out) + anion B(in) = anion A(in) + anion B(out).
amiloride-sensitive sodium channel activity
NA
sodium ion transmembrane transporter activity
Catalysis of the transfer of sodium ions (Na+) from one side of a membrane to the other.
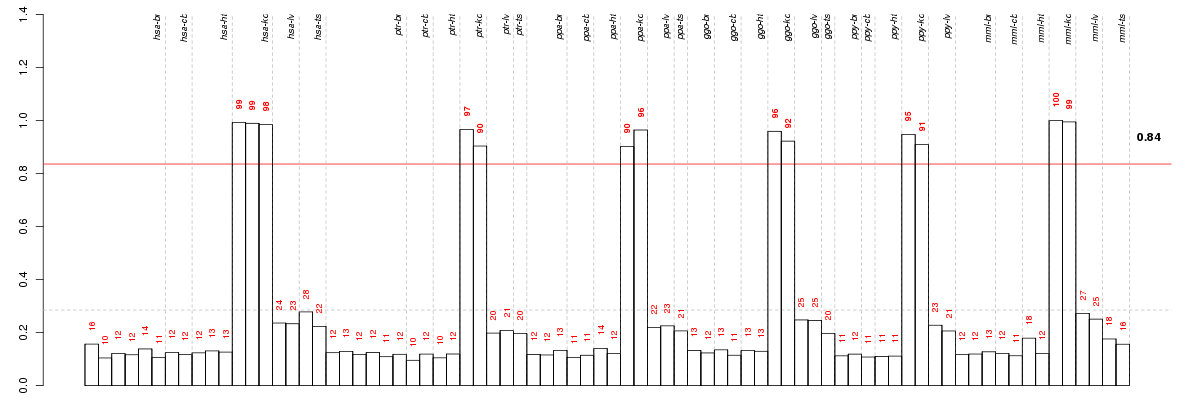
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04960 | 3.863e-04 | 0.5162 | 6 | 33 | Aldosterone-regulated sodium reabsorption |
| 04966 | 5.381e-04 | 0.3285 | 5 | 21 | Collecting duct acid secretion |
| 05110 | 8.382e-03 | 0.6257 | 5 | 40 | Vibrio cholerae infection |
| 05120 | 2.513e-02 | 0.829 | 5 | 53 | Epithelial cell signaling in Helicobacter pylori infection |
| 04670 | 4.157e-02 | 1.392 | 6 | 89 | Leukocyte transendothelial migration |
ABP1amiloride binding protein 1 (amine oxidase (copper-containing)) (ENSG00000002726), score: 0.85 ACPPacid phosphatase, prostate (ENSG00000014257), score: 0.93 ACSF2acyl-CoA synthetase family member 2 (ENSG00000167107), score: 0.79 ACY1aminoacylase 1 (ENSG00000243989), score: 0.74 ACY3aspartoacylase (aminocyclase) 3 (ENSG00000132744), score: 0.78 AGR2anterior gradient homolog 2 (Xenopus laevis) (ENSG00000106541), score: 0.77 AGXT2alanine--glyoxylate aminotransferase 2 (ENSG00000113492), score: 0.74 AP1M2adaptor-related protein complex 1, mu 2 subunit (ENSG00000129354), score: 0.87 AQP2aquaporin 2 (collecting duct) (ENSG00000167580), score: 0.99 AQP6aquaporin 6, kidney specific (ENSG00000086159), score: 0.96 ARHGAP24Rho GTPase activating protein 24 (ENSG00000138639), score: 0.75 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 1 ATP6V0D2ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 (ENSG00000147614), score: 0.96 ATP6V1B1ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 (ENSG00000116039), score: 0.96 ATP6V1G3ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 (ENSG00000151418), score: 0.98 B4GALNT2beta-1,4-N-acetyl-galactosaminyl transferase 2 (ENSG00000167080), score: 0.75 BARX2BARX homeobox 2 (ENSG00000043039), score: 0.76 BICC1bicaudal C homolog 1 (Drosophila) (ENSG00000122870), score: 0.8 BSNDBartter syndrome, infantile, with sensorineural deafness (Barttin) (ENSG00000162399), score: 0.97 BSPRYB-box and SPRY domain containing (ENSG00000119411), score: 0.74 C12orf59chromosome 12 open reading frame 59 (ENSG00000165685), score: 0.99 C19orf21chromosome 19 open reading frame 21 (ENSG00000099812), score: 0.94 C1orf106chromosome 1 open reading frame 106 (ENSG00000163362), score: 0.95 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.84 C1orf172chromosome 1 open reading frame 172 (ENSG00000175707), score: 0.77 C5orf32chromosome 5 open reading frame 32 (ENSG00000120306), score: 0.78 C8orf84chromosome 8 open reading frame 84 (ENSG00000164764), score: 0.8 C9orf71chromosome 9 open reading frame 71 (ENSG00000181778), score: 0.95 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.96 CALCRcalcitonin receptor (ENSG00000004948), score: 0.83 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.94 CDCP1CUB domain containing protein 1 (ENSG00000163814), score: 0.83 CDH16cadherin 16, KSP-cadherin (ENSG00000166589), score: 1 CDH3cadherin 3, type 1, P-cadherin (placental) (ENSG00000062038), score: 0.83 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (ENSG00000113361), score: 0.77 CDHR5cadherin-related family member 5 (ENSG00000099834), score: 0.74 CLCNKAchloride channel Ka (ENSG00000186510), score: 0.94 CLDN16claudin 16 (ENSG00000113946), score: 0.96 CLDN19claudin 19 (ENSG00000164007), score: 0.91 CLDN4claudin 4 (ENSG00000189143), score: 0.9 CLDN8claudin 8 (ENSG00000156284), score: 0.99 CLNKcytokine-dependent hematopoietic cell linker (ENSG00000109684), score: 0.93 CLRN3clarin 3 (ENSG00000180745), score: 0.79 COL4A3collagen, type IV, alpha 3 (Goodpasture antigen) (ENSG00000169031), score: 0.88 COL4A4collagen, type IV, alpha 4 (ENSG00000081052), score: 0.76 CUBNcubilin (intrinsic factor-cobalamin receptor) (ENSG00000107611), score: 0.96 CWH43cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) (ENSG00000109182), score: 0.85 CYP24A1cytochrome P450, family 24, subfamily A, polypeptide 1 (ENSG00000019186), score: 0.87 CYP27B1cytochrome P450, family 27, subfamily B, polypeptide 1 (ENSG00000111012), score: 0.9 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (ENSG00000153071), score: 0.8 DDCdopa decarboxylase (aromatic L-amino acid decarboxylase) (ENSG00000132437), score: 0.78 DEFB1defensin, beta 1 (ENSG00000164825), score: 0.78 DHDHdihydrodiol dehydrogenase (dimeric) (ENSG00000104808), score: 0.83 DMRT2doublesex and mab-3 related transcription factor 2 (ENSG00000173253), score: 0.94 DOCK8dedicator of cytokinesis 8 (ENSG00000107099), score: 0.79 DPEP1dipeptidase 1 (renal) (ENSG00000015413), score: 0.87 DPP4dipeptidyl-peptidase 4 (ENSG00000197635), score: 0.75 DUSP9dual specificity phosphatase 9 (ENSG00000130829), score: 0.88 EAF2ELL associated factor 2 (ENSG00000145088), score: 0.76 EGFepidermal growth factor (ENSG00000138798), score: 0.94 EHFets homologous factor (ENSG00000135373), score: 0.86 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.92 EPCAMepithelial cell adhesion molecule (ENSG00000119888), score: 0.88 EPS8L1EPS8-like 1 (ENSG00000131037), score: 0.86 ERP27endoplasmic reticulum protein 27 (ENSG00000139055), score: 0.75 ESRP1epithelial splicing regulatory protein 1 (ENSG00000104413), score: 0.97 EVPLenvoplakin (ENSG00000167880), score: 0.82 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.96 FAM151Afamily with sequence similarity 151, member A (ENSG00000162391), score: 0.76 FAM3Bfamily with sequence similarity 3, member B (ENSG00000183844), score: 0.74 FCAMRFc receptor, IgA, IgM, high affinity (ENSG00000162897), score: 0.84 FMO1flavin containing monooxygenase 1 (ENSG00000010932), score: 0.92 FOLR1folate receptor 1 (adult) (ENSG00000110195), score: 0.8 FOXI1forkhead box I1 (ENSG00000168269), score: 1 FRMD1FERM domain containing 1 (ENSG00000153303), score: 0.84 FXYD2FXYD domain containing ion transport regulator 2 (ENSG00000137731), score: 0.91 GALNT14UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) (ENSG00000158089), score: 0.76 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.91 GCM1glial cells missing homolog 1 (Drosophila) (ENSG00000137270), score: 0.75 GDF15growth differentiation factor 15 (ENSG00000130513), score: 0.8 GGT6gamma-glutamyltransferase 6 (ENSG00000167741), score: 0.92 GHRHRgrowth hormone releasing hormone receptor (ENSG00000106128), score: 0.75 GIPC2GIPC PDZ domain containing family, member 2 (ENSG00000137960), score: 0.8 GP2glycoprotein 2 (zymogen granule membrane) (ENSG00000169347), score: 0.91 GPR110G protein-coupled receptor 110 (ENSG00000153292), score: 0.98 GPR160G protein-coupled receptor 160 (ENSG00000173890), score: 0.77 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.8 GRB7growth factor receptor-bound protein 7 (ENSG00000141738), score: 0.75 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.98 GYLTL1Bglycosyltransferase-like 1B (ENSG00000165905), score: 0.82 HEPACAM2HEPACAM family member 2 (ENSG00000188175), score: 0.91 HHLA2HERV-H LTR-associating 2 (ENSG00000114455), score: 0.75 HMX2H6 family homeobox 2 (ENSG00000188816), score: 0.94 HNF1BHNF1 homeobox B (ENSG00000108753), score: 0.83 HOXA10homeobox A10 (ENSG00000153807), score: 0.99 HOXA11homeobox A11 (ENSG00000005073), score: 0.81 HOXA3homeobox A3 (ENSG00000105997), score: 0.84 HOXA5homeobox A5 (ENSG00000106004), score: 0.92 HOXA7homeobox A7 (ENSG00000122592), score: 0.88 HOXA9homeobox A9 (ENSG00000078399), score: 1 HOXB5homeobox B5 (ENSG00000120075), score: 0.95 HOXB6homeobox B6 (ENSG00000108511), score: 0.91 HOXB7homeobox B7 (ENSG00000120087), score: 0.95 HOXC10homeobox C10 (ENSG00000180818), score: 0.98 HOXC6homeobox C6 (ENSG00000197757), score: 0.92 HOXC8homeobox C8 (ENSG00000037965), score: 0.93 HOXC9homeobox C9 (ENSG00000180806), score: 0.92 HOXD1homeobox D1 (ENSG00000128645), score: 0.78 HOXD10homeobox D10 (ENSG00000128710), score: 0.99 HOXD3homeobox D3 (ENSG00000128652), score: 0.93 HOXD4homeobox D4 (ENSG00000170166), score: 0.95 HOXD8homeobox D8 (ENSG00000175879), score: 0.85 HOXD9homeobox D9 (ENSG00000128709), score: 0.93 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.94 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.95 INSRRinsulin receptor-related receptor (ENSG00000027644), score: 0.92 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 1 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (ENSG00000153822), score: 0.84 KCNK5potassium channel, subfamily K, member 5 (ENSG00000164626), score: 0.76 KIF12kinesin family member 12 (ENSG00000136883), score: 0.8 KLklotho (ENSG00000133116), score: 0.98 KLHDC7Akelch domain containing 7A (ENSG00000179023), score: 0.87 KRT7keratin 7 (ENSG00000135480), score: 0.84 LAD1ladinin 1 (ENSG00000159166), score: 0.75 LGALS2lectin, galactoside-binding, soluble, 2 (ENSG00000100079), score: 0.91 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.84 LRRC19leucine rich repeat containing 19 (ENSG00000184434), score: 0.92 MACC1metastasis associated in colon cancer 1 (ENSG00000183742), score: 0.87 MCCD1mitochondrial coiled-coil domain 1 (ENSG00000204511), score: 0.94 MIOXmyo-inositol oxygenase (ENSG00000100253), score: 0.96 MMEmembrane metallo-endopeptidase (ENSG00000196549), score: 0.82 MMP7matrix metallopeptidase 7 (matrilysin, uterine) (ENSG00000137673), score: 0.83 MUC1mucin 1, cell surface associated (ENSG00000185499), score: 0.85 MYO3Bmyosin IIIB (ENSG00000071909), score: 0.77 NHSNance-Horan syndrome (congenital cataracts and dental anomalies) (ENSG00000188158), score: 0.76 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.96 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 0.98 NPNTnephronectin (ENSG00000168743), score: 0.78 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.77 NUAK2NUAK family, SNF1-like kinase, 2 (ENSG00000163545), score: 0.75 OXGR1oxoglutarate (alpha-ketoglutarate) receptor 1 (ENSG00000165621), score: 0.76 PAPPApregnancy-associated plasma protein A, pappalysin 1 (ENSG00000182752), score: 0.84 PAQR5progestin and adipoQ receptor family member V (ENSG00000137819), score: 0.8 PARD6Bpar-6 partitioning defective 6 homolog beta (C. elegans) (ENSG00000124171), score: 0.82 PAX2paired box 2 (ENSG00000075891), score: 0.96 PAX8paired box 8 (ENSG00000125618), score: 0.97 PDZK1IP1PDZK1 interacting protein 1 (ENSG00000162366), score: 0.96 PIGRpolymeric immunoglobulin receptor (ENSG00000162896), score: 0.75 PKHD1polycystic kidney and hepatic disease 1 (autosomal recessive) (ENSG00000170927), score: 0.92 PLA2G4Fphospholipase A2, group IVF (ENSG00000168907), score: 0.96 PLA2R1phospholipase A2 receptor 1, 180kDa (ENSG00000153246), score: 0.77 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.79 PPP1R14Dprotein phosphatase 1, regulatory (inhibitor) subunit 14D (ENSG00000166143), score: 0.83 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (ENSG00000111725), score: 0.82 PRLRprolactin receptor (ENSG00000113494), score: 0.77 PROM2prominin 2 (ENSG00000155066), score: 0.91 PRR15proline rich 15 (ENSG00000176532), score: 0.88 PRR15Lproline rich 15-like (ENSG00000167183), score: 0.84 PRSS8protease, serine, 8 (ENSG00000052344), score: 0.86 PTGER3prostaglandin E receptor 3 (subtype EP3) (ENSG00000050628), score: 0.82 PTH1Rparathyroid hormone 1 receptor (ENSG00000160801), score: 0.83 RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.92 RAB25RAB25, member RAS oncogene family (ENSG00000132698), score: 0.95 RAB7L1RAB7, member RAS oncogene family-like 1 (ENSG00000117280), score: 0.79 RASSF10Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 (ENSG00000189431), score: 0.79 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.76 RENrenin (ENSG00000143839), score: 0.86 RGL3ral guanine nucleotide dissociation stimulator-like 3 (ENSG00000205517), score: 0.78 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.93 S100A2S100 calcium binding protein A2 (ENSG00000196754), score: 0.87 SCINscinderin (ENSG00000006747), score: 0.91 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.91 SCNN1Bsodium channel, nonvoltage-gated 1, beta (ENSG00000168447), score: 0.9 SCNN1Gsodium channel, nonvoltage-gated 1, gamma (ENSG00000166828), score: 0.89 SCTRsecretin receptor (ENSG00000080293), score: 0.82 SIM1single-minded homolog 1 (Drosophila) (ENSG00000112246), score: 0.93 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.85 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.94 SLC12A1solute carrier family 12 (sodium/potassium/chloride transporters), member 1 (ENSG00000074803), score: 0.96 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (ENSG00000070915), score: 0.99 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.89 SLC13A3solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 (ENSG00000158296), score: 0.86 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.81 SLC16A4solute carrier family 16, member 4 (monocarboxylic acid transporter 5) (ENSG00000168679), score: 0.93 SLC16A9solute carrier family 16, member 9 (monocarboxylic acid transporter 9) (ENSG00000165449), score: 0.87 SLC17A1solute carrier family 17 (sodium phosphate), member 1 (ENSG00000124568), score: 0.76 SLC17A3solute carrier family 17 (sodium phosphate), member 3 (ENSG00000124564), score: 0.8 SLC22A11solute carrier family 22 (organic anion/urate transporter), member 11 (ENSG00000168065), score: 0.96 SLC22A12solute carrier family 22 (organic anion/urate transporter), member 12 (ENSG00000197891), score: 1 SLC22A13solute carrier family 22 (organic anion transporter), member 13 (ENSG00000172940), score: 0.85 SLC22A2solute carrier family 22 (organic cation transporter), member 2 (ENSG00000112499), score: 0.97 SLC22A5solute carrier family 22 (organic cation/carnitine transporter), member 5 (ENSG00000197375), score: 0.76 SLC22A6solute carrier family 22 (organic anion transporter), member 6 (ENSG00000197901), score: 0.93 SLC22A8solute carrier family 22 (organic anion transporter), member 8 (ENSG00000149452), score: 0.97 SLC23A3solute carrier family 23 (nucleobase transporters), member 3 (ENSG00000213901), score: 0.76 SLC26A7solute carrier family 26, member 7 (ENSG00000147606), score: 0.95 SLC30A2solute carrier family 30 (zinc transporter), member 2 (ENSG00000158014), score: 0.75 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.99 SLC34A3solute carrier family 34 (sodium phosphate), member 3 (ENSG00000198569), score: 0.97 SLC35F2solute carrier family 35, member F2 (ENSG00000110660), score: 0.77 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (ENSG00000138079), score: 0.75 SLC3A2solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 (ENSG00000168003), score: 0.76 SLC44A3solute carrier family 44, member 3 (ENSG00000143036), score: 0.79 SLC44A4solute carrier family 44, member 4 (ENSG00000204385), score: 0.97 SLC47A1solute carrier family 47, member 1 (ENSG00000142494), score: 0.78 SLC47A2solute carrier family 47, member 2 (ENSG00000180638), score: 0.98 SLC4A1solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) (ENSG00000004939), score: 0.98 SLC4A9solute carrier family 4, sodium bicarbonate cotransporter, member 9 (ENSG00000113073), score: 0.98 SLC5A10solute carrier family 5 (sodium/glucose cotransporter), member 10 (ENSG00000154025), score: 0.88 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.98 SLC5A2solute carrier family 5 (sodium/glucose cotransporter), member 2 (ENSG00000140675), score: 0.82 SLC5A8solute carrier family 5 (iodide transporter), member 8 (ENSG00000139357), score: 0.77 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.98 SLC7A13solute carrier family 7, (cationic amino acid transporter, y+ system) member 13 (ENSG00000164893), score: 0.83 SLC7A8solute carrier family 7 (amino acid transporter, L-type), member 8 (ENSG00000092068), score: 0.83 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 0.8 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.91 SLCO4C1solute carrier organic anion transporter family, member 4C1 (ENSG00000173930), score: 0.89 SMTNL2smoothelin-like 2 (ENSG00000188176), score: 0.79 SOSTsclerostin (ENSG00000167941), score: 0.8 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.81 STK32Bserine/threonine kinase 32B (ENSG00000152953), score: 0.78 STRA6stimulated by retinoic acid gene 6 homolog (mouse) (ENSG00000137868), score: 0.94 SULT1C2sulfotransferase family, cytosolic, 1C, member 2 (ENSG00000198203), score: 0.88 SUSD2sushi domain containing 2 (ENSG00000099994), score: 0.82 TACSTD2tumor-associated calcium signal transducer 2 (ENSG00000184292), score: 0.94 TBX2T-box 2 (ENSG00000121068), score: 0.79 TCN2transcobalamin II (ENSG00000185339), score: 0.75 TFAP2Atranscription factor AP-2 alpha (activating enhancer binding protein 2 alpha) (ENSG00000137203), score: 0.79 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.75 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.99 TFECtranscription factor EC (ENSG00000105967), score: 0.87 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.99 TMC4transmembrane channel-like 4 (ENSG00000167608), score: 0.81 TMEM106Atransmembrane protein 106A (ENSG00000184988), score: 0.78 TMEM139transmembrane protein 139 (ENSG00000178826), score: 0.82 TMEM171transmembrane protein 171 (ENSG00000157111), score: 0.91 TMEM174transmembrane protein 174 (ENSG00000164325), score: 0.99 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.88 TMEM72transmembrane protein 72 (ENSG00000187783), score: 0.96 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.85 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.82 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.8 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.89 TSPAN1tetraspanin 1 (ENSG00000117472), score: 0.84 TSPAN33tetraspanin 33 (ENSG00000158457), score: 0.76 TTC22tetratricopeptide repeat domain 22 (ENSG00000006555), score: 0.82 UMODuromodulin (ENSG00000169344), score: 1 UNC5CLunc-5 homolog C (C. elegans)-like (ENSG00000124602), score: 0.81 USH1CUsher syndrome 1C (autosomal recessive, severe) (ENSG00000006611), score: 0.87 VAV3vav 3 guanine nucleotide exchange factor (ENSG00000134215), score: 0.8 VDRvitamin D (1,25- dihydroxyvitamin D3) receptor (ENSG00000111424), score: 0.96 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.79 WFDC2WAP four-disulfide core domain 2 (ENSG00000101443), score: 0.83 WNK4WNK lysine deficient protein kinase 4 (ENSG00000126562), score: 0.96 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.88
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ppy_kd_f_ca1 | ppy | kd | f | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ppy_kd_m_ca1 | ppy | kd | m | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |