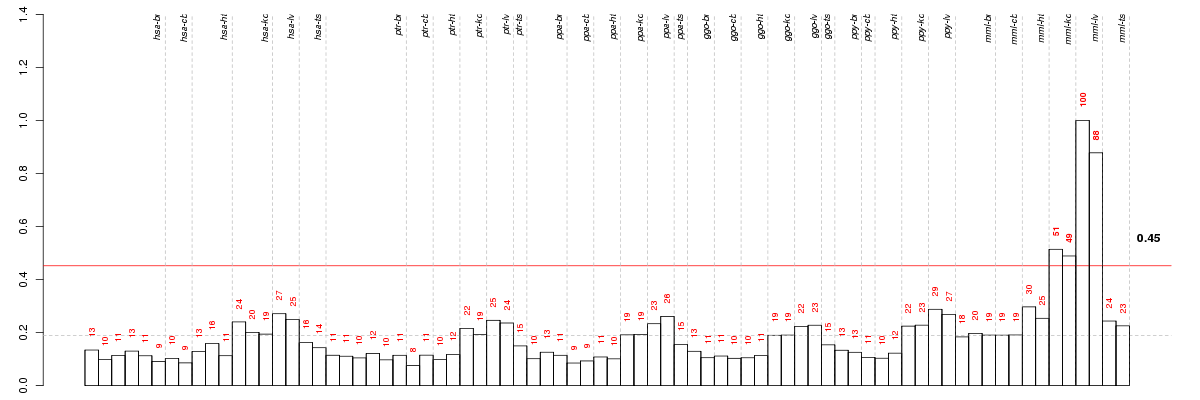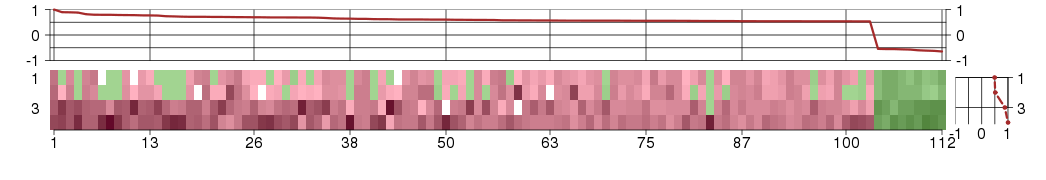

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to radiation
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an electromagnetic radiation stimulus. Electromagnetic radiation is a propagating wave in space with electric and magnetic components. These components oscillate at right angles to each other and to the direction of propagation.
response to light stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a light stimulus, electromagnetic radiation of wavelengths classified as infrared, visible or ultraviolet light.
detection of external stimulus
The series of events in which an external stimulus is received by a cell and converted into a molecular signal.
detection of abiotic stimulus
The series of events in which an (non-living) abiotic stimulus is received by a cell and converted into a molecular signal.
detection of light stimulus
The series of events in which a light stimulus (in the form of photons) is received and converted into a molecular signal.
detection of visible light
The series of events in which a visible light stimulus is received by a cell and converted into a molecular signal. A visible light stimulus is electromagnetic radiation that can be perceived visually by an organism; for organisms lacking a visual system, this can be defined as light with a wavelength within the range 380 to 780 nm.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to abiotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an abiotic (non-living) stimulus.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
detection of stimulus
The series of events in which a stimulus is received by a cell or organism and converted into a molecular signal.
all
NA
detection of external stimulus
The series of events in which an external stimulus is received by a cell and converted into a molecular signal.
detection of abiotic stimulus
The series of events in which an (non-living) abiotic stimulus is received by a cell and converted into a molecular signal.
detection of light stimulus
The series of events in which a light stimulus (in the form of photons) is received and converted into a molecular signal.
detection of light stimulus
The series of events in which a light stimulus (in the form of photons) is received and converted into a molecular signal.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
photoreceptor outer segment
The outer segment of a vertebrate photoreceptor that contains discs of photoreceptive membranes.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cilium
A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
nonmotile primary cilium
An immotile primary cilium that may be missing the central pair of microtubules, or the central pair of microtubules and outer dynein arms. Some primary cilia also have altered arrangements of outer microtubules (fewer than nine and/or not always present as doublets). Nonmotile primary cilia typically function as sensory organelles that concentrate and organize sensory signaling molecules.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
photoreceptor outer segment membrane
The membrane surrounding the outer segment of a vertebrate photoreceptor.

A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.69 ABCA4ATP-binding cassette, sub-family A (ABC1), member 4 (ENSG00000198691), score: 0.7 ABP1amiloride binding protein 1 (amine oxidase (copper-containing)) (ENSG00000002726), score: 0.56 ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.56 ADAM12ADAM metallopeptidase domain 12 (ENSG00000148848), score: 0.89 AFPalpha-fetoprotein (ENSG00000081051), score: 0.82 AIM1Labsent in melanoma 1-like (ENSG00000176092), score: 0.69 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.63 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.65 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: 0.7 ANO7anoctamin 7 (ENSG00000146205), score: 0.56 APOA4apolipoprotein A-IV (ENSG00000110244), score: 0.69 AQP8aquaporin 8 (ENSG00000103375), score: 0.56 ARAP1ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (ENSG00000186635), score: 0.54 ARSBarylsulfatase B (ENSG00000113273), score: 0.56 ART4ADP-ribosyltransferase 4 (Dombrock blood group) (ENSG00000111339), score: 0.61 B3GNT7UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 (ENSG00000156966), score: 0.68 BIKBCL2-interacting killer (apoptosis-inducing) (ENSG00000100290), score: 0.54 BIRC7baculoviral IAP repeat-containing 7 (ENSG00000101197), score: 0.61 C17orf87chromosome 17 open reading frame 87 (ENSG00000161929), score: 0.56 C1orf177chromosome 1 open reading frame 177 (ENSG00000162398), score: 0.57 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.6 C22orf15chromosome 22 open reading frame 15 (ENSG00000169314), score: 0.56 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.59 C3orf26chromosome 3 open reading frame 26 (ENSG00000184220), score: -0.55 C9orf152chromosome 9 open reading frame 152 (ENSG00000188959), score: 0.79 CAMTA1calmodulin binding transcription activator 1 (ENSG00000171735), score: -0.56 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.54 CCL23chemokine (C-C motif) ligand 23 (ENSG00000167236), score: 0.71 CCL24chemokine (C-C motif) ligand 24 (ENSG00000106178), score: 0.57 CD1ECD1e molecule (ENSG00000158488), score: 0.6 CD33CD33 molecule (ENSG00000105383), score: 0.54 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.61 CDX2caudal type homeobox 2 (ENSG00000165556), score: 0.64 CEACAM16carcinoembryonic antigen-related cell adhesion molecule 16 (ENSG00000213892), score: 0.53 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.56 CRYBA2crystallin, beta A2 (ENSG00000163499), score: 0.77 CXCR2chemokine (C-X-C motif) receptor 2 (ENSG00000180871), score: 0.61 CXCR3chemokine (C-X-C motif) receptor 3 (ENSG00000186810), score: 0.6 CYFIP1cytoplasmic FMR1 interacting protein 1 (ENSG00000068793), score: 0.55 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.58 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.71 DSC3desmocollin 3 (ENSG00000134762), score: 0.78 EMR1egf-like module containing, mucin-like, hormone receptor-like 1 (ENSG00000174837), score: 0.59 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.77 FAM189Bfamily with sequence similarity 189, member B (ENSG00000160767), score: -0.65 FGD2FYVE, RhoGEF and PH domain containing 2 (ENSG00000146192), score: 0.55 FLJ35220hypothetical protein FLJ35220 (ENSG00000173818), score: -0.54 FOXP3forkhead box P3 (ENSG00000049768), score: 0.7 FPR3formyl peptide receptor 3 (ENSG00000187474), score: 0.57 GAS8growth arrest-specific 8 (ENSG00000141013), score: 0.56 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.54 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.72 GNAT1guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 (ENSG00000114349), score: 0.68 GNAT2guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 (ENSG00000134183), score: 0.54 GPR152G protein-coupled receptor 152 (ENSG00000175514), score: 0.55 GUCA2Bguanylate cyclase activator 2B (uroguanylin) (ENSG00000044012), score: 0.53 GYLTL1Bglycosyltransferase-like 1B (ENSG00000165905), score: 0.54 HRASLS2HRAS-like suppressor 2 (ENSG00000133328), score: 0.8 IFIT1Binterferon-induced protein with tetratricopeptide repeats 1B (ENSG00000204010), score: 0.54 IGFL3IGF-like family member 3 (ENSG00000188624), score: 0.56 IL2interleukin 2 (ENSG00000109471), score: 0.55 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.57 JMJD4jumonji domain containing 4 (ENSG00000081692), score: 0.53 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.71 KRT28keratin 28 (ENSG00000173908), score: 0.89 LGMNlegumain (ENSG00000100600), score: 0.55 LIPMlipase, family member M (ENSG00000173239), score: 0.62 LOC100127905family with sequence similarity 165, member B pseudogene (ENSG00000198738), score: 0.54 LRRC8Eleucine rich repeat containing 8 family, member E (ENSG00000171017), score: 0.55 LYPD3LY6/PLAUR domain containing 3 (ENSG00000124466), score: 0.69 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.71 MPV17LMPV17 mitochondrial membrane protein-like (ENSG00000156968), score: 0.59 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.56 NUMBnumb homolog (Drosophila) (ENSG00000133961), score: 0.56 NYXnyctalopin (ENSG00000188937), score: 0.6 OR10H3olfactory receptor, family 10, subfamily H, member 3 (ENSG00000171936), score: 0.69 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.56 PAEPprogestagen-associated endometrial protein (ENSG00000122133), score: 0.69 PARP15poly (ADP-ribose) polymerase family, member 15 (ENSG00000173200), score: 0.54 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.71 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.62 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.64 PRHOXNBparahox cluster neighbor (ENSG00000183463), score: 0.88 PROL1proline rich, lacrimal 1 (ENSG00000171199), score: 0.59 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.71 PSKH2protein serine kinase H2 (ENSG00000147613), score: 0.76 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.56 RGS13regulator of G-protein signaling 13 (ENSG00000127074), score: 0.79 RHBDD3rhomboid domain containing 3 (ENSG00000100263), score: 0.54 RNASE1ribonuclease, RNase A family, 1 (pancreatic) (ENSG00000129538), score: -0.62 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.57 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.61 SERPINA12serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (ENSG00000165953), score: 0.73 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.58 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.54 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.6 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.56 SMPDL3Asphingomyelin phosphodiesterase, acid-like 3A (ENSG00000172594), score: 0.57 SOAT2sterol O-acyltransferase 2 (ENSG00000167780), score: 0.62 SRD5A3steroid 5 alpha-reductase 3 (ENSG00000128039), score: 0.56 TBX19T-box 19 (ENSG00000143178), score: 0.63 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.55 TLR6toll-like receptor 6 (ENSG00000174130), score: 0.67 TNNtenascin N (ENSG00000120332), score: 0.54 TRPM5transient receptor potential cation channel, subfamily M, member 5 (ENSG00000070985), score: 0.79 UCMAupper zone of growth plate and cartilage matrix associated (ENSG00000165623), score: 0.57 VWCEvon Willebrand factor C and EGF domains (ENSG00000167992), score: 0.6 ZBTB3zinc finger and BTB domain containing 3 (ENSG00000185670), score: 0.54
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |