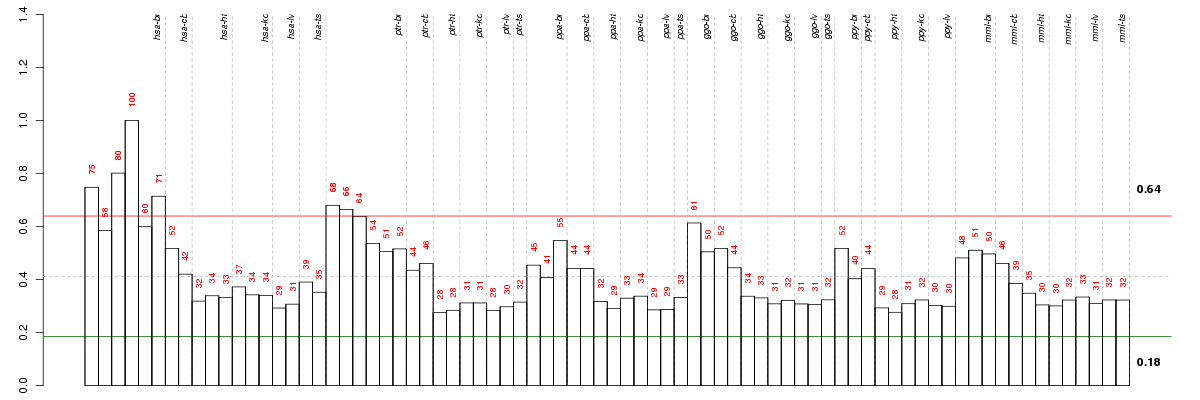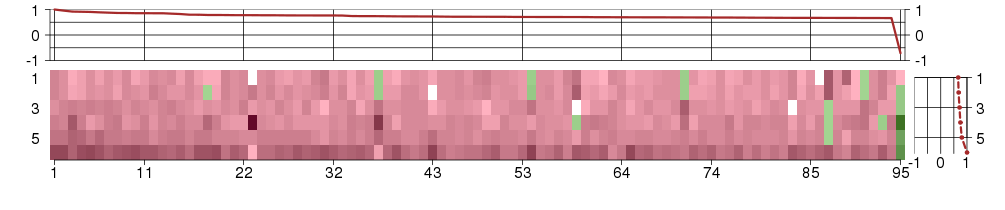

Under-expression is coded with green,
over-expression with red color.

cell fate specification
The process involved in the specification of cell identity. Once specification has taken place, a cell will be committed to differentiate down a specific pathway if left in its normal environment.
cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
spinal cord development
The process whose specific outcome is the progression of the spinal cord over time, from its formation to the mature structure. The spinal cord primarily conducts sensory and motor nerve impulses between the brain and the peripheral nervous tissues.
cell differentiation in spinal cord
The process whereby relatively unspecialized cells acquire specialized structural and/or functional features that characterize the cells of the spinal cord. Differentiation includes the processes involved in commitment of a cell to a specific fate.
spinal cord oligodendrocyte cell differentiation
The process whereby neuroepithelial cells in the neural tube acquire specialized structural and/or functional features of oligodendrocytes. Oligodendrocytes are non-neuronal cells. The primary function of oligodendrocytes is the myelination of nerve axons in the central nervous system. Differentiation includes the processes involved in commitment of a cell to a specific fate.
spinal cord oligodendrocyte cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into an oligodendrocyte in an environment that is neutral with respect to the developmental pathway.
oligodendrocyte cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into an oligodendrocyte in an environment that is neutral with respect to the developmental pathway. Upon specification, the cell fate can be reversed.
oligodendrocyte cell fate commitment
The process whereby the developmental fate of a cell becomes restricted such that it will develop into an oligodendrocyte.
glial cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into a glial cell in an environment that is neutral with respect to the developmental pathway. Upon specification, the cell fate can be reversed.
glial cell fate commitment
The process whereby the developmental fate of a cell becomes restricted such that it will develop into a glial cell.
neurogenesis
Generation of cells within the nervous system.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
gliogenesis
The process by which glial cells are generated. This includes the production of glial progenitors and their differentiation into mature glia.
cell fate commitment
The commitment of cells to specific cell fates and their capacity to differentiate into particular kinds of cells. Positional information is established through protein signals that emanate from a localized source within a cell (the initial one-cell zygote) or within a developmental field.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
all
NA
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cell fate commitment
The commitment of cells to specific cell fates and their capacity to differentiate into particular kinds of cells. Positional information is established through protein signals that emanate from a localized source within a cell (the initial one-cell zygote) or within a developmental field.
cell fate specification
The process involved in the specification of cell identity. Once specification has taken place, a cell will be committed to differentiate down a specific pathway if left in its normal environment.
cell differentiation in spinal cord
The process whereby relatively unspecialized cells acquire specialized structural and/or functional features that characterize the cells of the spinal cord. Differentiation includes the processes involved in commitment of a cell to a specific fate.
glial cell fate commitment
The process whereby the developmental fate of a cell becomes restricted such that it will develop into a glial cell.
glial cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into a glial cell in an environment that is neutral with respect to the developmental pathway. Upon specification, the cell fate can be reversed.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
neurogenesis
Generation of cells within the nervous system.
spinal cord development
The process whose specific outcome is the progression of the spinal cord over time, from its formation to the mature structure. The spinal cord primarily conducts sensory and motor nerve impulses between the brain and the peripheral nervous tissues.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
spinal cord oligodendrocyte cell differentiation
The process whereby neuroepithelial cells in the neural tube acquire specialized structural and/or functional features of oligodendrocytes. Oligodendrocytes are non-neuronal cells. The primary function of oligodendrocytes is the myelination of nerve axons in the central nervous system. Differentiation includes the processes involved in commitment of a cell to a specific fate.
oligodendrocyte cell fate commitment
The process whereby the developmental fate of a cell becomes restricted such that it will develop into an oligodendrocyte.
spinal cord oligodendrocyte cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into an oligodendrocyte in an environment that is neutral with respect to the developmental pathway.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
oligodendrocyte cell fate specification
The process whereby a cell becomes capable of differentiating autonomously into an oligodendrocyte in an environment that is neutral with respect to the developmental pathway. Upon specification, the cell fate can be reversed.


ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (ENSG00000107331), score: 0.72 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.72 ATP10BATPase, class V, type 10B (ENSG00000118322), score: 0.77 C11orf9chromosome 11 open reading frame 9 (ENSG00000124920), score: 0.77 C1orf198chromosome 1 open reading frame 198 (ENSG00000119280), score: 0.77 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.72 CAPN3calpain 3, (p94) (ENSG00000092529), score: 0.72 CAPN9calpain 9 (ENSG00000135773), score: 0.77 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: -0.71 CD22CD22 molecule (ENSG00000012124), score: 0.96 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.86 CHADLchondroadherin-like (ENSG00000100399), score: 0.82 CLCA4chloride channel accessory 4 (ENSG00000016602), score: 0.9 CLDN11claudin 11 (ENSG00000013297), score: 0.67 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.77 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.79 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.69 CMTM5CKLF-like MARVEL transmembrane domain containing 5 (ENSG00000166091), score: 0.72 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (ENSG00000173786), score: 0.85 CPNE2copine II (ENSG00000140848), score: 0.74 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.76 DMBT1deleted in malignant brain tumors 1 (ENSG00000187908), score: 0.73 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.74 EFSembryonal Fyn-associated substrate (ENSG00000100842), score: 0.69 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.8 EML2echinoderm microtubule associated protein like 2 (ENSG00000125746), score: 0.71 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.73 ERMNermin, ERM-like protein (ENSG00000136541), score: 0.67 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.78 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.8 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.68 FAM108B1family with sequence similarity 108, member B1 (ENSG00000107362), score: 0.69 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.86 FAM125Bfamily with sequence similarity 125, member B (ENSG00000196814), score: 0.67 FFAR1free fatty acid receptor 1 (ENSG00000126266), score: 0.7 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.7 GAB2GRB2-associated binding protein 2 (ENSG00000033327), score: 0.71 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (ENSG00000139629), score: 0.89 GIPC1GIPC PDZ domain containing family, member 1 (ENSG00000123159), score: 0.66 GLDNgliomedin (ENSG00000186417), score: 0.78 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (ENSG00000170775), score: 0.71 GPR78G protein-coupled receptor 78 (ENSG00000155269), score: 0.78 GREM1gremlin 1 (ENSG00000166923), score: 0.71 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.69 IFNA2interferon, alpha 2 (ENSG00000188379), score: 0.67 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.85 KREMEN2kringle containing transmembrane protein 2 (ENSG00000131650), score: 0.92 KRT83keratin 83 (ENSG00000170523), score: 0.67 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.86 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (ENSG00000107902), score: 0.67 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.67 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.74 MAGmyelin associated glycoprotein (ENSG00000105695), score: 0.72 MARCKSL1MARCKS-like 1 (ENSG00000175130), score: 0.74 MATN1matrilin 1, cartilage matrix protein (ENSG00000162510), score: 0.67 MKRN3makorin ring finger protein 3 (ENSG00000179455), score: 0.71 MOGmyelin oligodendrocyte glycoprotein (ENSG00000204655), score: 0.7 MYO9Bmyosin IXB (ENSG00000099331), score: 0.69 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 1 NKX2-2NK2 homeobox 2 (ENSG00000125820), score: 0.73 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.91 OLIG2oligodendrocyte lineage transcription factor 2 (ENSG00000205927), score: 0.68 OPALINoligodendrocytic myelin paranodal and inner loop protein (ENSG00000197430), score: 0.7 P2RX7purinergic receptor P2X, ligand-gated ion channel, 7 (ENSG00000089041), score: 0.67 PAQR6progestin and adipoQ receptor family member VI (ENSG00000160781), score: 0.68 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.77 PLEKHG3pleckstrin homology domain containing, family G (with RhoGef domain) member 3 (ENSG00000126822), score: 0.68 PLXNB3plexin B3 (ENSG00000198753), score: 0.69 PPP1R14Aprotein phosphatase 1, regulatory (inhibitor) subunit 14A (ENSG00000167641), score: 0.71 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.74 PRKCQprotein kinase C, theta (ENSG00000065675), score: 0.71 PRR18proline rich 18 (ENSG00000176381), score: 0.88 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.69 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.78 PTPRHprotein tyrosine phosphatase, receptor type, H (ENSG00000080031), score: 0.78 PXKPX domain containing serine/threonine kinase (ENSG00000168297), score: 0.69 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.68 RHOGras homolog gene family, member G (rho G) (ENSG00000177105), score: 0.7 ROM1retinal outer segment membrane protein 1 (ENSG00000149489), score: 0.7 SEMA4Dsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D (ENSG00000187764), score: 0.67 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.69 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.68 SH3TC2SH3 domain and tetratricopeptide repeats 2 (ENSG00000169247), score: 0.73 SIRT2sirtuin 2 (ENSG00000068903), score: 0.86 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.77 SOX10SRY (sex determining region Y)-box 10 (ENSG00000100146), score: 0.71 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.73 TBCBtubulin folding cofactor B (ENSG00000105254), score: 0.68 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.77 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.7 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.84 TMEM151Atransmembrane protein 151A (ENSG00000179292), score: 0.67 TP53INP2tumor protein p53 inducible nuclear protein 2 (ENSG00000078804), score: 0.77 UGT8UDP glycosyltransferase 8 (ENSG00000174607), score: 0.69 ZFP57zinc finger protein 57 homolog (mouse) (ENSG00000204644), score: 0.67
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |