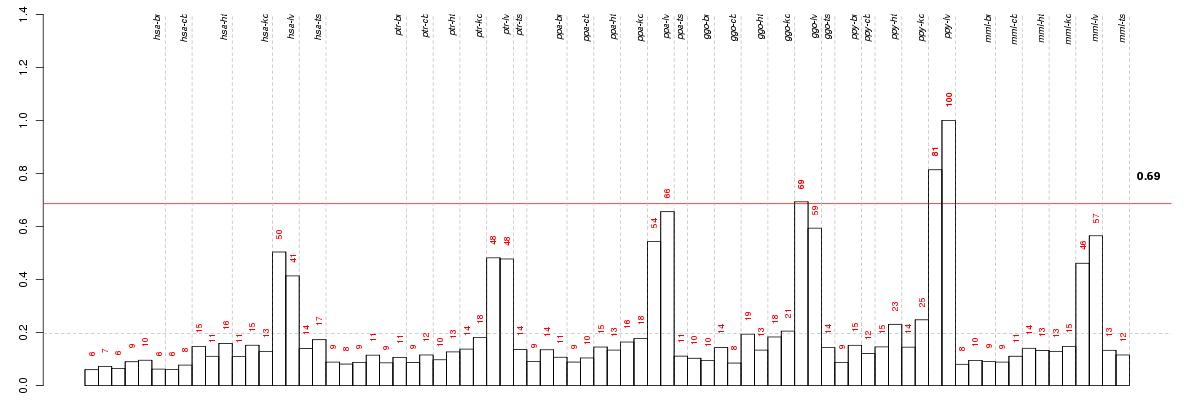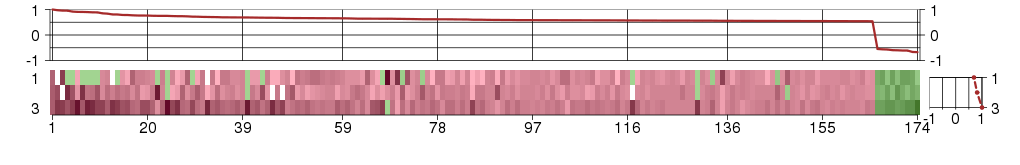

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cell activation
A change in the morphology or behavior of a cell resulting from exposure to an activating factor such as a cellular or soluble ligand.
cytokine production
The appearance of a cytokine due to biosynthesis or secretion following a cellular stimulus, resulting in an increase in its intracellular or extracellular levels.
regulation of cytokine production
Any process that modulates the frequency, rate, or extent of production of a cytokine.
positive regulation of cytokine production
Any process that activates or increases the frequency, rate or extent of production of a cytokine.
cell killing
Any process in an organism that results in the killing of its own cells or those of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
leukocyte mediated cytotoxicity
The directed killing of a target cell by a leukocyte.
immune effector process
Any process of the immune system that occurs as part of an immune response.
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
myeloid leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a myeloid leukocyte.
neutrophil mediated immunity
Any process involved in the carrying out of an immune response by a neutrophil.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
acute inflammatory response
Inflammation which comprises a rapid, short-lived, relatively uniform response to acute injury or antigenic challenge and is characterized by accumulations of fluid, plasma proteins, and granulocytic leukocytes. An acute inflammatory response occurs within a matter of minutes or hours, and either resolves within a few days or becomes a chronic inflammatory response.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
defense response to bacterium
Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism.
defense response to fungus
Reactions triggered in response to the presence of a fungus that act to protect the cell or organism.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
humoral immune response
An immune response mediated through a body fluid.
cellular defense response
A defense response that is mediated by cells.
growth
The increase in size or mass of an entire organism, a part of an organism or a cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
intracellular protein kinase cascade
A series of reactions that occur within the cell, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
JAK-STAT cascade
Any process by which STAT proteins (Signal Transducers and Activators of Transcription) are activated by members of the JAK (janus activated kinase) family of tyrosine kinases, following the binding of cytokines to their cognate receptor. Once activated, STATs dimerize and translocate to the nucleus and modulate the expression of target genes.
hemostasis
The stopping of bleeding (loss of body fluid) or the arrest of the circulation to an organ or part.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
cell death
A biological process that results in permanent cessation of all vital functions of a cell.
cytolysis
The rupture of cell membranes and the loss of cytoplasm.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
response to bacterium
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium.
response to fungus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a fungus.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cell death
Any process that modulates the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
death
A permanent cessation of all vital functions: the end of life; can be applied to a whole organism or to a part of an organism.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
signal transmission via phosphorylation event
The process whereby a signal is conveyed via the transfer of one or more phosphate groups.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
regulation of signaling process
Any process that modulates the frequency, rate or extent of a signaling process.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
positive regulation of signaling process
Any process that activates, maintains or increases the frequency, rate or extent of a signaling process.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
symbiosis, encompassing mutualism through parasitism
An interaction between two organisms living together in more or less intimate association. The term host is usually used for the larger (macro) of the two members of a symbiosis. The smaller (micro) member is called the symbiont organism. Microscopic symbionts are often referred to as endosymbionts. The various forms of symbiosis include parasitism, in which the association is disadvantageous or destructive to one of the organisms; mutualism, in which the association is advantageous, or often necessary to one or both and not harmful to either; and commensalism, in which one member of the association benefits while the other is not affected. However, mutualism, parasitism, and commensalism are often not discrete categories of interactions and should rather be perceived as a continuum of interaction ranging from parasitism to mutualism. In fact, the direction of a symbiotic interaction can change during the lifetime of the symbionts due to developmental changes as well as changes in the biotic/abiotic environment in which the interaction occurs.
killing of cells of another organism
Any process in an organism that results in the killing of cells of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
regulation of growth of symbiont in host
Any process by which the symbiont regulates the increase in its size or mass within the cells or tissues of the host organism. This may (but not necessarily) include a filamentous growth form, and also can include secretion of proteases and lipases to break down. The host is defined as the larger of the organisms involved in the symbiotic interaction.
negative regulation of growth of symbiont in host
Any process by which the symbiont stops, prevents or reduces its increase in size or mass within the cells or tissues of the host organism. The host is defined as the larger of the organisms involved in the symbiotic interaction.
regulation of signaling pathway
Any process that modulates the frequency, rate or extent of a signaling pathway.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
regulation of growth
Any process that modulates the frequency, rate or extent of the growth of all or part of an organism so that it occurs at its proper speed, either globally or in a specific part of the organism's development.
wound healing
The series of events that restore integrity to a damaged tissue, following an injury.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
regulation of cytolysis
Any process that modulates the frequency, rate or extent of the rupture of cell membranes and the loss of cytoplasm.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
regulation of multi-organism process
Any process that modulates the frequency, rate or extent of a multi-organism process, a process by which an organism has an effect on another organism of the same or different species.
negative regulation of multi-organism process
Any process that stops, prevents or reduces the frequency, rate or extent of a multi-organism process, a process by which an organism has an effect on another organism of the same or different species.
regulation of symbiosis, encompassing mutualism through parasitism
Any process that modulates the frequency, rate or extent of symbiosis, an interaction between two organisms living together in more or less intimate association.
growth involved in symbiotic interaction
The increase in size or mass of an organism occurring when the organism is in a symbiotic interaction.
growth of symbiont involved in interaction with host
The increase in size or mass of an organism, occurring in, on or near the exterior of its host organism. The host is defined as the larger of the organisms involved in a symbiotic interaction.
growth of symbiont in host
The increase in size or mass of an organism, occurring within the cells or tissues of the host organism. This may (but not necessarily) include a filamentous growth form, and also can include secretion of proteases and lipases to break down host tissue. The host is defined as the larger of the organisms involved in a symbiotic interaction.
modulation of growth of symbiont involved in interaction with host
Any process that modulates the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
negative regulation of growth of symbiont involved in interaction with host
Any process that stops, prevents or reduces the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
interspecies interaction between organisms
Any process by which an organism has an effect on an organism of a different species.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
leukocyte activation
A change in morphology and behavior of a leukocyte resulting from exposure to a specific antigen, mitogen, cytokine, cellular ligand, or soluble factor.
negative regulation of cytolysis
Any process that stops, prevents or reduces the frequency, rate or extent of cytolysis.
negative regulation of growth
Any process that stops, prevents or reduces the rate or extent of growth, the increase in size or mass of all or part of an organism.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
coagulation
The process by which a fluid solution, or part of it, changes into a solid or semisolid mass.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
interaction with symbiont
An interaction between two organisms living together in more or less intimate association. The term symbiont is used for the smaller (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
modification of morphology or physiology of other organism involved in symbiotic interaction
The process by which an organism effects a change in the structure or processes of a second organism, where the two organisms are in a symbiotic interaction.
disruption of cells of other organism involved in symbiotic interaction
A process by which an organism has a negative effect on the functioning of the second organism's cells, where the two organisms are in a symbiotic interaction.
modification by host of symbiont morphology or physiology
The process by which an organism effects a change in the structure or processes of a symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
disruption by host of symbiont cells
Any process by which an organism has a negative effect on the functioning of the symbiont's cells. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing by host of symbiont cells
Any process mediated by an organism that results in the death of cells in the symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing of cells in other organism involved in symbiotic interaction
Any process mediated by an organism that results in the death of cells in a second organism, where the two organisms are in a symbiotic interaction.
negative regulation of cell death
Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
neutrophil mediated cytotoxicity
The directed killing of a target cell by a neutrophil.
neutrophil mediated killing of symbiont cell
The directed killing of a symbiont target cell by a neutrophil.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cell death
A biological process that results in permanent cessation of all vital functions of a cell.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
negative regulation of growth
Any process that stops, prevents or reduces the rate or extent of growth, the increase in size or mass of all or part of an organism.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
regulation of signaling process
Any process that modulates the frequency, rate or extent of a signaling process.
regulation of growth
Any process that modulates the frequency, rate or extent of the growth of all or part of an organism so that it occurs at its proper speed, either globally or in a specific part of the organism's development.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
killing of cells of another organism
Any process in an organism that results in the killing of cells of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions.
regulation of multi-organism process
Any process that modulates the frequency, rate or extent of a multi-organism process, a process by which an organism has an effect on another organism of the same or different species.
negative regulation of multi-organism process
Any process that stops, prevents or reduces the frequency, rate or extent of a multi-organism process, a process by which an organism has an effect on another organism of the same or different species.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
leukocyte mediated cytotoxicity
The directed killing of a target cell by a leukocyte.
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of growth
Any process that stops, prevents or reduces the rate or extent of growth, the increase in size or mass of all or part of an organism.
leukocyte activation
A change in morphology and behavior of a leukocyte resulting from exposure to a specific antigen, mitogen, cytokine, cellular ligand, or soluble factor.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
negative regulation of cell death
Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cell death
Any process that modulates the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
regulation of signaling pathway
Any process that modulates the frequency, rate or extent of a signaling pathway.
regulation of cytokine production
Any process that modulates the frequency, rate, or extent of production of a cytokine.
positive regulation of multicellular organismal process
Any process that activates or increases the frequency, rate or extent of an organismal process, any of the processes pertinent to the function of an organism above the cellular level; includes the integrated processes of tissues and organs.
positive regulation of cytokine production
Any process that activates or increases the frequency, rate or extent of production of a cytokine.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of growth of symbiont involved in interaction with host
Any process that stops, prevents or reduces the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
negative regulation of multi-organism process
Any process that stops, prevents or reduces the frequency, rate or extent of a multi-organism process, a process by which an organism has an effect on another organism of the same or different species.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
regulation of body fluid levels
Any process that modulates the levels of body fluids.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
negative regulation of growth of symbiont involved in interaction with host
Any process that stops, prevents or reduces the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
modulation of growth of symbiont involved in interaction with host
Any process that modulates the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
negative regulation of growth of symbiont involved in interaction with host
Any process that stops, prevents or reduces the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
negative regulation of growth of symbiont in host
Any process by which the symbiont stops, prevents or reduces its increase in size or mass within the cells or tissues of the host organism. The host is defined as the larger of the organisms involved in the symbiotic interaction.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of cell communication
Any process that increases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
positive regulation of signaling pathway
Any process that activates or increases the frequency, rate or extent of a signaling pathway.
regulation of cytolysis
Any process that modulates the frequency, rate or extent of the rupture of cell membranes and the loss of cytoplasm.
negative regulation of cell death
Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death.
negative regulation of cytolysis
Any process that stops, prevents or reduces the frequency, rate or extent of cytolysis.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
positive regulation of signal transduction
Any process that activates or increases the frequency, rate or extent of signal transduction.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
positive regulation of cytokine production
Any process that activates or increases the frequency, rate or extent of production of a cytokine.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
regulation of growth of symbiont in host
Any process by which the symbiont regulates the increase in its size or mass within the cells or tissues of the host organism. This may (but not necessarily) include a filamentous growth form, and also can include secretion of proteases and lipases to break down. The host is defined as the larger of the organisms involved in the symbiotic interaction.
modulation of growth of symbiont involved in interaction with host
Any process that modulates the frequency, rate or extent of the increase in size or mass of an organism occurring in, on or near the exterior of its host organism.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
defense response to bacterium
Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism.
defense response to fungus
Reactions triggered in response to the presence of a fungus that act to protect the cell or organism.
regulation of symbiosis, encompassing mutualism through parasitism
Any process that modulates the frequency, rate or extent of symbiosis, an interaction between two organisms living together in more or less intimate association.
growth involved in symbiotic interaction
The increase in size or mass of an organism occurring when the organism is in a symbiotic interaction.
interaction with symbiont
An interaction between two organisms living together in more or less intimate association. The term symbiont is used for the smaller (macro) of the two members of a symbiosis; the various forms of symbiosis include parasitism, commensalism and mutualism.
modification of morphology or physiology of other organism involved in symbiotic interaction
The process by which an organism effects a change in the structure or processes of a second organism, where the two organisms are in a symbiotic interaction.
modification by host of symbiont morphology or physiology
The process by which an organism effects a change in the structure or processes of a symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
neutrophil mediated killing of symbiont cell
The directed killing of a symbiont target cell by a neutrophil.
regulation of growth of symbiont in host
Any process by which the symbiont regulates the increase in its size or mass within the cells or tissues of the host organism. This may (but not necessarily) include a filamentous growth form, and also can include secretion of proteases and lipases to break down. The host is defined as the larger of the organisms involved in the symbiotic interaction.
negative regulation of growth of symbiont in host
Any process by which the symbiont stops, prevents or reduces its increase in size or mass within the cells or tissues of the host organism. The host is defined as the larger of the organisms involved in the symbiotic interaction.
negative regulation of cytolysis
Any process that stops, prevents or reduces the frequency, rate or extent of cytolysis.
intracellular protein kinase cascade
A series of reactions that occur within the cell, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
blood coagulation
The sequential process by which the multiple coagulation factors of the blood interact, ultimately resulting in the formation of an insoluble fibrin clot; it may be divided into three stages: stage 1, the formation of intrinsic and extrinsic prothrombin converting principle; stage 2, the formation of thrombin; stage 3, the formation of stable fibrin polymers.
disruption by host of symbiont cells
Any process by which an organism has a negative effect on the functioning of the symbiont's cells. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
killing of cells in other organism involved in symbiotic interaction
Any process mediated by an organism that results in the death of cells in a second organism, where the two organisms are in a symbiotic interaction.
neutrophil mediated cytotoxicity
The directed killing of a target cell by a neutrophil.
killing by host of symbiont cells
Any process mediated by an organism that results in the death of cells in the symbiont organism. The symbiont is defined as the smaller of the organisms involved in a symbiotic interaction.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
stored secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell and upon stimulation, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
platelet alpha granule
A secretory organelle found in blood platelets, which is unique in that it exhibits further compartmentalization and acquires its protein content via two distinct mechanisms: (1) biosynthesis predominantly at the megakaryocyte (MK) level (with some vestigial platelet synthesis) (e.g. platelet factor 4) and (2) endocytosis and pinocytosis at both the MK and circulating platelet levels (e.g. fibrinogen (Fg) and IgG).
platelet alpha granule lumen
The volume enclosed by the membrane of the platelet alpha granule.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
vesicle lumen
The volume enclosed by the membrane or protein that forms a vesicle.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
vesicle lumen
The volume enclosed by the membrane or protein that forms a vesicle.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic membrane-bounded vesicle lumen
The volume enclosed by the membrane of a cytoplasmic membrane-bounded vesicle.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
platelet alpha granule lumen
The volume enclosed by the membrane of the platelet alpha granule.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
serine-type endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain.
serine-type peptidase activity
Catalysis of the hydrolysis of peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
peptidase activity
Catalysis of the hydrolysis of a peptide bond. A peptide bond is a covalent bond formed when the carbon atom from the carboxyl group of one amino acid shares electrons with the nitrogen atom from the amino group of a second amino acid.
enzyme inhibitor activity
Stops, prevents or reduces the activity of an enzyme.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
carbohydrate binding
Interacting selectively and non-covalently with any carbohydrate, any of a group of organic compounds based of the general formula Cx(H2O)y.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
sugar binding
Interacting selectively and non-covalently with any mono-, di- or trisaccharide carbohydrate.
lipid binding
Interacting selectively and non-covalently with a lipid.
peptidase activity, acting on L-amino acid peptides
Catalysis of the hydrolysis of peptide bonds formed between L-amino acids.
endopeptidase regulator activity
Modulates the activity of a peptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
serine hydrolase activity
Catalysis of the hydrolysis of a substrate by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
enzyme regulator activity
Modulates the activity of an enzyme.
cell surface binding
Interacting selectively and non-covalently with any component on the surface of a cell.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
peptidase regulator activity
Modulates the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
all
NA
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
serine-type peptidase activity
Catalysis of the hydrolysis of peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
serine-type endopeptidase activity
Catalysis of the hydrolysis of internal, alpha-peptide bonds in a polypeptide chain by a catalytic mechanism that involves a catalytic triad consisting of a serine nucleophile that is activated by a proton relay involving an acidic residue (e.g. aspartate or glutamate) and a basic residue (usually histidine).
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 5.943e-04 | 3.768 | 14 | 198 | Cytokine-cytokine receptor interaction |
| 04610 | 1.207e-03 | 0.9515 | 7 | 50 | Complement and coagulation cascades |
| 05322 | 4.709e-02 | 0.9896 | 5 | 52 | Systemic lupus erythematosus |
ABHD12Babhydrolase domain containing 12B (ENSG00000131969), score: 0.55 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.55 ANKRD11ankyrin repeat domain 11 (ENSG00000167522), score: -0.56 AOAHacyloxyacyl hydrolase (neutrophil) (ENSG00000136250), score: 0.56 APCSamyloid P component, serum (ENSG00000132703), score: 0.56 APOFapolipoprotein F (ENSG00000175336), score: 0.62 AQP8aquaporin 8 (ENSG00000103375), score: 0.68 ARNTaryl hydrocarbon receptor nuclear translocator (ENSG00000143437), score: 0.61 ARR3arrestin 3, retinal (X-arrestin) (ENSG00000120500), score: 0.59 BAATbile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) (ENSG00000136881), score: 0.54 BMP10bone morphogenetic protein 10 (ENSG00000163217), score: 0.65 BPIL1bactericidal/permeability-increasing protein-like 1 (ENSG00000078898), score: 0.9 BPIL3bactericidal/permeability-increasing protein-like 3 (ENSG00000167104), score: 0.75 BST1bone marrow stromal cell antigen 1 (ENSG00000109743), score: 0.56 C16orf54chromosome 16 open reading frame 54 (ENSG00000185905), score: 0.64 C17orf48chromosome 17 open reading frame 48 (ENSG00000170222), score: 0.54 C17orf67chromosome 17 open reading frame 67 (ENSG00000214226), score: 0.56 C20orf70chromosome 20 open reading frame 70 (ENSG00000131050), score: 0.89 C2orf66chromosome 2 open reading frame 66 (ENSG00000187944), score: 0.81 C5complement component 5 (ENSG00000106804), score: 0.55 C8Bcomplement component 8, beta polypeptide (ENSG00000021852), score: 0.57 C9complement component 9 (ENSG00000113600), score: 0.56 CAMPcathelicidin antimicrobial peptide (ENSG00000164047), score: 0.68 CARD17caspase recruitment domain family, member 17 (ENSG00000137752), score: 0.59 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.57 CCL20chemokine (C-C motif) ligand 20 (ENSG00000115009), score: 0.64 CCRL2chemokine (C-C motif) receptor-like 2 (ENSG00000121797), score: 0.57 CD209CD209 molecule (ENSG00000090659), score: 0.76 CD300LFCD300 molecule-like family member f (ENSG00000186074), score: 0.58 CD5CD5 molecule (ENSG00000110448), score: 0.61 CD69CD69 molecule (ENSG00000110848), score: 0.58 CEACAM8carcinoembryonic antigen-related cell adhesion molecule 8 (ENSG00000124469), score: 0.68 CHIT1chitinase 1 (chitotriosidase) (ENSG00000133063), score: 0.63 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (ENSG00000140835), score: 0.77 CLEC4GC-type lectin domain family 4, member G (ENSG00000182566), score: 0.56 CLEC5AC-type lectin domain family 5, member A (ENSG00000090269), score: 0.64 COLEC10collectin sub-family member 10 (C-type lectin) (ENSG00000184374), score: 0.58 CPceruloplasmin (ferroxidase) (ENSG00000047457), score: 0.55 CPB2carboxypeptidase B2 (plasma) (ENSG00000080618), score: 0.55 CRISP3cysteine-rich secretory protein 3 (ENSG00000096006), score: 0.74 CSF2colony stimulating factor 2 (granulocyte-macrophage) (ENSG00000164400), score: 0.69 CST7cystatin F (leukocystatin) (ENSG00000077984), score: 0.55 CTSGcathepsin G (ENSG00000100448), score: 0.67 CTSKcathepsin K (ENSG00000143387), score: 0.62 CTSWcathepsin W (ENSG00000172543), score: 0.59 CXCR1chemokine (C-X-C motif) receptor 1 (ENSG00000163464), score: 0.58 CXCR6chemokine (C-X-C motif) receptor 6 (ENSG00000172215), score: 0.58 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.55 DHODHdihydroorotate dehydrogenase (ENSG00000102967), score: 0.57 DNAH11dynein, axonemal, heavy chain 11 (ENSG00000105877), score: 0.67 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.58 ELANEelastase, neutrophil expressed (ENSG00000197561), score: 0.56 EVI2Becotropic viral integration site 2B (ENSG00000185862), score: 0.54 F9coagulation factor IX (ENSG00000101981), score: 0.55 FAM83Afamily with sequence similarity 83, member A (ENSG00000147689), score: 1 FCARFc fragment of IgA, receptor for (ENSG00000186431), score: 0.7 FCN2ficolin (collagen/fibrinogen domain containing lectin) 2 (hucolin) (ENSG00000160339), score: 0.56 FGBfibrinogen beta chain (ENSG00000171564), score: 0.56 FGF21fibroblast growth factor 21 (ENSG00000105550), score: 0.69 FGF23fibroblast growth factor 23 (ENSG00000118972), score: 0.57 FGGfibrinogen gamma chain (ENSG00000171557), score: 0.56 FSTfollistatin (ENSG00000134363), score: 0.58 GNGT1guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 (ENSG00000127928), score: 0.8 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.67 GPR15G protein-coupled receptor 15 (ENSG00000154165), score: 0.63 GPR97G protein-coupled receptor 97 (ENSG00000182885), score: 0.76 GPRC5DG protein-coupled receptor, family C, group 5, member D (ENSG00000111291), score: 0.71 GPX2glutathione peroxidase 2 (gastrointestinal) (ENSG00000176153), score: 0.57 GRAP2GRB2-related adaptor protein 2 (ENSG00000100351), score: 0.68 GRHL1grainyhead-like 1 (Drosophila) (ENSG00000134317), score: 0.66 GTF2IRD1GTF2I repeat domain containing 1 (ENSG00000006704), score: -0.61 GZMKgranzyme K (granzyme 3; tryptase II) (ENSG00000113088), score: 0.71 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.65 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (ENSG00000019991), score: 0.58 HIST1H1Bhistone cluster 1, H1b (ENSG00000184357), score: 0.75 HPS3Hermansky-Pudlak syndrome 3 (ENSG00000163755), score: 0.62 HPSEheparanase (ENSG00000173083), score: 0.7 HPXhemopexin (ENSG00000110169), score: 0.56 HSH2Dhematopoietic SH2 domain containing (ENSG00000196684), score: 0.58 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.64 IFNAR1interferon (alpha, beta and omega) receptor 1 (ENSG00000142166), score: 0.67 IL18RAPinterleukin 18 receptor accessory protein (ENSG00000115607), score: 0.58 IL1RAPinterleukin 1 receptor accessory protein (ENSG00000196083), score: 0.59 IL1RNinterleukin 1 receptor antagonist (ENSG00000136689), score: 0.63 IL7Rinterleukin 7 receptor (ENSG00000168685), score: 0.66 ITIH3inter-alpha (globulin) inhibitor H3 (ENSG00000162267), score: 0.54 ITKIL2-inducible T-cell kinase (ENSG00000113263), score: 0.66 KLRC1killer cell lectin-like receptor subfamily C, member 1 (ENSG00000134545), score: 0.58 KLRD1killer cell lectin-like receptor subfamily D, member 1 (ENSG00000134539), score: 0.62 KRT27keratin 27 (ENSG00000171446), score: 0.96 LAX1lymphocyte transmembrane adaptor 1 (ENSG00000122188), score: 0.73 LBPlipopolysaccharide binding protein (ENSG00000129988), score: 0.57 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (ENSG00000187116), score: 0.66 LOC100134052similar to ubiquitin associated and SH3 domain containing, A (ENSG00000160185), score: 0.66 LONP2lon peptidase 2, peroxisomal (ENSG00000102910), score: 0.61 LRG1leucine-rich alpha-2-glycoprotein 1 (ENSG00000171236), score: 0.54 MED17mediator complex subunit 17 (ENSG00000042429), score: 0.6 MLANAmelan-A (ENSG00000120215), score: 0.97 MMP8matrix metallopeptidase 8 (neutrophil collagenase) (ENSG00000118113), score: 0.91 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (ENSG00000100985), score: 0.58 MSMBmicroseminoprotein, beta- (ENSG00000138294), score: 0.92 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (ENSG00000163738), score: 0.74 MXD1MAX dimerization protein 1 (ENSG00000059728), score: 0.64 MYBPHmyosin binding protein H (ENSG00000133055), score: 0.62 MYO1Fmyosin IF (ENSG00000142347), score: 0.57 MYO7Amyosin VIIA (ENSG00000137474), score: 0.56 NCR1natural cytotoxicity triggering receptor 1 (ENSG00000189430), score: 0.69 NF2neurofibromin 2 (merlin) (ENSG00000186575), score: -0.61 NFE2nuclear factor (erythroid-derived 2), 45kDa (ENSG00000123405), score: 0.68 NFIL3nuclear factor, interleukin 3 regulated (ENSG00000165030), score: 0.54 NLRP12NLR family, pyrin domain containing 12 (ENSG00000142405), score: 0.85 NR5A2nuclear receptor subfamily 5, group A, member 2 (ENSG00000116833), score: 0.57 OFD1oral-facial-digital syndrome 1 (ENSG00000046651), score: 0.55 OIT3oncoprotein induced transcript 3 (ENSG00000138315), score: 0.61 OR10G2olfactory receptor, family 10, subfamily G, member 2 (ENSG00000169202), score: 0.64 ORM1orosomucoid 1 (ENSG00000229314), score: 0.58 OSMoncostatin M (ENSG00000099985), score: 0.66 PDCD1LG2programmed cell death 1 ligand 2 (ENSG00000197646), score: 0.58 PF4platelet factor 4 (ENSG00000163737), score: 0.65 PGM3phosphoglucomutase 3 (ENSG00000013375), score: 0.59 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (ENSG00000105647), score: -0.55 PLEKpleckstrin (ENSG00000115956), score: 0.58 PPBPpro-platelet basic protein (chemokine (C-X-C motif) ligand 7) (ENSG00000163736), score: 0.75 PRAM1PML-RARA regulated adaptor molecule 1 (ENSG00000133246), score: 0.57 PRDM1PR domain containing 1, with ZNF domain (ENSG00000057657), score: 0.66 PRLprolactin (ENSG00000172179), score: 0.9 PRSS53protease, serine, 53 (ENSG00000151006), score: 0.57 PTPN22protein tyrosine phosphatase, non-receptor type 22 (lymphoid) (ENSG00000134242), score: 0.55 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.55 PYHIN1pyrin and HIN domain family, member 1 (ENSG00000163564), score: 0.56 RASGEF1BRasGEF domain family, member 1B (ENSG00000138670), score: 0.54 RASGRP4RAS guanyl releasing protein 4 (ENSG00000171777), score: 0.57 RFWD2ring finger and WD repeat domain 2 (ENSG00000143207), score: 0.56 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.62 S100A12S100 calcium binding protein A12 (ENSG00000163221), score: 0.69 S100PS100 calcium binding protein P (ENSG00000163993), score: 0.57 S1PR4sphingosine-1-phosphate receptor 4 (ENSG00000125910), score: 0.62 SAMSN1SAM domain, SH3 domain and nuclear localization signals 1 (ENSG00000155307), score: 0.54 SCGB3A2secretoglobin, family 3A, member 2 (ENSG00000164265), score: 0.89 SELLselectin L (ENSG00000188404), score: 0.66 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.58 SERPINA11serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 (ENSG00000186910), score: 0.55 SERPINB10serpin peptidase inhibitor, clade B (ovalbumin), member 10 (ENSG00000242550), score: 0.96 SIGLEC14sialic acid binding Ig-like lectin 14 (ENSG00000105501), score: 0.76 SLAMF7SLAM family member 7 (ENSG00000026751), score: 0.66 SLC20A1solute carrier family 20 (phosphate transporter), member 1 (ENSG00000144136), score: 0.68 SLC28A3solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 (ENSG00000197506), score: 0.69 SMG5Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) (ENSG00000198952), score: -0.59 SNX20sorting nexin 20 (ENSG00000167208), score: 0.64 SPINK1serine peptidase inhibitor, Kazal type 1 (ENSG00000164266), score: 0.6 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.56 SRP19signal recognition particle 19kDa (ENSG00000153037), score: 0.58 STAB2stabilin 2 (ENSG00000136011), score: 0.57 SULT2A1sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (ENSG00000105398), score: 0.59 TATtyrosine aminotransferase (ENSG00000198650), score: 0.55 TBC1D10CTBC1 domain family, member 10C (ENSG00000175463), score: 0.54 TBC1D14TBC1 domain family, member 14 (ENSG00000132405), score: -0.67 TCN1transcobalamin I (vitamin B12 binding protein, R binder family) (ENSG00000134827), score: 0.6 TDO2tryptophan 2,3-dioxygenase (ENSG00000151790), score: 0.57 TFF1trefoil factor 1 (ENSG00000160182), score: 0.72 TIGITT cell immunoreceptor with Ig and ITIM domains (ENSG00000181847), score: 0.84 TMC3transmembrane channel-like 3 (ENSG00000188869), score: 0.72 TMEM149transmembrane protein 149 (ENSG00000126246), score: 0.55 TMEM92transmembrane protein 92 (ENSG00000167105), score: 0.55 TNFRSF9tumor necrosis factor receptor superfamily, member 9 (ENSG00000049249), score: 0.74 TOB1transducer of ERBB2, 1 (ENSG00000141232), score: 0.64 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.79 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.55 USP2ubiquitin specific peptidase 2 (ENSG00000036672), score: -0.6 VNN2vanin 2 (ENSG00000112303), score: 0.75 WISP1WNT1 inducible signaling pathway protein 1 (ENSG00000104415), score: 0.69 ZNF498zinc finger protein 498 (ENSG00000197037), score: 0.78
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ppy_lv_m_ca1 | ppy | lv | m | _ |
| ppy_lv_f_ca1 | ppy | lv | f | _ |