Difference between revisions of "SNP minor allele frequencies and their functional implication"
Sbprm2021 2 (talk | contribs) |
Sbprm2021 2 (talk | contribs) |
||
| Line 26: | Line 26: | ||
====Methods==== | ====Methods==== | ||
| − | SNPs frequencies were found on the gnomAD v2.1.1 data set containing data from 125,748 exomes and 15,708 whole genomes, all mapped to the GRCh37/hg19. Data were extracted (577’000 entries) using the terminal and python. The information selected from the data were the chromosome, the gene name, the ID SNP number, the position, the minor allele frequency (contained in a minimum of 10 individuals), the codon change, the amino acid substitution, and the type of variant. For the analysis, R studio was used in order to do the statistical tests (Fisher tests) as well as the plots. Pantherdb was used to study biological functions of the synonymous | + | SNPs frequencies were found on the gnomAD v2.1.1 data set containing data from 125,748 exomes and 15,708 whole genomes, all mapped to the GRCh37/hg19. Data were extracted (577’000 entries) using the terminal and python. The information selected from the data were the chromosome, the gene name, the ID SNP number, the position, the minor allele frequency (contained in a minimum of 10 individuals), the codon change, the amino acid substitution, and the type of variant. For the analysis, R studio was used in order to do the statistical tests (Fisher tests) as well as the plots. Pantherdb was used to study the biological functions of the genes which have 100% synonymous mutations. |
The project was separated into 3 parts: | The project was separated into 3 parts: | ||
| Line 50: | Line 50: | ||
The results showed a significant difference (p-value < 2.2e-16) with a Fisher-test comparing the two extremes bin. | The results showed a significant difference (p-value < 2.2e-16) with a Fisher-test comparing the two extremes bin. | ||
| − | The results were the following, SNPs at high frequency tend to have more synonymous mutations compared to low SNPs frequencies where they tend to be missense. This suggests that the mutations which do not change the amino acid are | + | The results were the following, SNPs at high frequency tend to have more synonymous mutations compared to low SNPs frequencies where they tend to be missense. This suggests that the mutations which do not change the amino acid (synonymous mutations) are less under a negative selective pressure than missense mutations. Therefore, they can spread more easily into the population and tend to be present at a higher proportion for high SNPs frequency. It is nevertheless necessary to remain attentive: we cannot talk about positive selection because as we said before we used minor allele frequency. Most likely SNPs at high frequency are more neutral and SNPs at low frequency are maybe more under the negative selection. |
| − | As we had the mutated position of the codons, we were interested | + | As we had the mutated position of the codons, we were interested in the distribution of the SNPs according to their frequencies by separating the groups with this new variable. To do this, we extracted the minor allele frequencies separated by the position of the codon change. At a high allele frequency, there is a bigger proportion of synonymous mutations. The type of mutation mostly depends on the mutated position of the codon. Therefore, we wanted to see if our previous result was consistent with the position of the codons that are mutated. |
Here we plotted the SNPs frequencies on the x-axis and the percentage of the positions mutated within the codons on the y axis. We can see that the more a SNP is frequent in the population, the more it tends to have a mutation in the third position of the codon. This was expected because this is consistent with the result (part I SNPs), where we saw that at high SNP frequency, the proportion of synonymous mutations is higher. There are fewer chances to change the amino acid if the third position of the codon is mutated. On the contrary, if the first or the second position of a codon is mutated, it has more chances to change the amino acid encoded. | Here we plotted the SNPs frequencies on the x-axis and the percentage of the positions mutated within the codons on the y axis. We can see that the more a SNP is frequent in the population, the more it tends to have a mutation in the third position of the codon. This was expected because this is consistent with the result (part I SNPs), where we saw that at high SNP frequency, the proportion of synonymous mutations is higher. There are fewer chances to change the amino acid if the third position of the codon is mutated. On the contrary, if the first or the second position of a codon is mutated, it has more chances to change the amino acid encoded. | ||
| Line 71: | Line 71: | ||
As you can see, for each function indicated in the list on the left, there is a higher percentage for the genes we submitted than for the genes of the reference list. | As you can see, for each function indicated in the list on the left, there is a higher percentage for the genes we submitted than for the genes of the reference list. | ||
| − | This suggests that the genes we submitted are not randomly involved in these functions. However, we need to be careful. These results showed that there is a correlation between the function of the gene and the frequency of synonymous mutations, but this is not causation. In other words, the function of the gene | + | This suggests that the genes we submitted are not randomly involved in these functions. However, we need to be careful. These results showed that there is a correlation between the function of the gene and the frequency of synonymous mutations, but this is not causation. In other words, the function of the gene does not allow to infer about the frequency of synonymous mutations it may have. |
| Line 80: | Line 80: | ||
There are only 54 genes that encode tRNA anticodons in the human genome. The 10 remaining codons without complementary tRNA anticodon can be recognized due to the wobble position and the edited tRNA anticodon. | There are only 54 genes that encode tRNA anticodons in the human genome. The 10 remaining codons without complementary tRNA anticodon can be recognized due to the wobble position and the edited tRNA anticodon. | ||
| − | To continue, we extracted the tRNA relative abundance associated | + | To continue, we extracted the tRNA relative abundance associated with each codon (for the original and the mutated). Then, we did the absolute difference between the tRNA abundance for the original codon and for the mutated one. The difference gave us the delta abundance and we compared it with the associated frequency of the SNP. |
Revision as of 14:08, 2 July 2021
- Project name: Frequencies of codon changing SNPs in a human population
- Tutor: Anneke Brümmer (anneke.brummer_AT_unil.ch)
- Slides: File:BPTRM Anneke 2021.pdf
- Report: [1]
Contents
Frequencies of codon changing SNPs in a human population
Aurélien Pirat, Mélissa Monn, Alicia Fauquex
Supervisor: Anneke Brümmer
Introduction
The genetic code is the system of correspondence allowing to translate a sequence of nucleic acids into protein. A triplet of nucleotides (or codon) designates a specific amino acid. As there are 4 nucleotides, we can have 64 different codons. In proteins, only 20 different amino acids are found. The 20 amino acid side chains are classified into 14 groups according to their hydrophobicity, polarity, size, charge, and potential for side-chain hydrogen bonding (1). Several codons can lead to the same amino acid; therefore, we say that the genetic code is redundant.
An anticodon is a group of three nucleotides that are in the structure of tRNAs, which plays an important role in mRNA translation during protein biosynthesis. This triplet of nucleotides located in a single-stranded loop can specifically pair with the codon complementary sequence present on the mRNA strand. More precisely, the tRNA carries at its 3' end the esterified amino acid corresponding to its anticodon. The pairing codon-anticodon takes place in the ribosome, the latter can add the required amino acid to the protein being synthesized.
Single nucleotide polymorphism (called SNPs) is the variation of a single base pair of the genome between individuals of the same species. This variation happens in almost one in a thousand base pairs in the human genome. Common SNPs must be located at a specific point in the genome and occurs in more than 1% of the population to be characterized as a SNP. Most of the SNPs used in this project have a lower frequency than 1%. They can be found in coding regions of genes (exon), non-coding regions of genes but also in intergenic regions, between genes. SNPs can be synonymous (not causing a change in the amino acid) or nonsynonymous (when the amino acid is altered). In our analysis, we focused on coding regions. In this case, SNPs will not necessarily modify the amino acid sequence of the protein produced thanks to the redundancy of the genetic code. However, SNPs in synonymous changes can have an effect on the protein folding, on protein abundances, or moreover on protein stability.
Methods
SNPs frequencies were found on the gnomAD v2.1.1 data set containing data from 125,748 exomes and 15,708 whole genomes, all mapped to the GRCh37/hg19. Data were extracted (577’000 entries) using the terminal and python. The information selected from the data were the chromosome, the gene name, the ID SNP number, the position, the minor allele frequency (contained in a minimum of 10 individuals), the codon change, the amino acid substitution, and the type of variant. For the analysis, R studio was used in order to do the statistical tests (Fisher tests) as well as the plots. Pantherdb was used to study the biological functions of the genes which have 100% synonymous mutations.
The project was separated into 3 parts:
- the allele frequency
- codon changes
- tRNA relative abundance
Part I : synonymous and non-synonymous SNPs variants
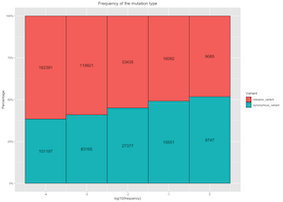
We were wondering if there was a difference in the SNP frequency spectrum for synonymous and missense variants. To visualize it, we plotted minor allele frequencies vs the percentage of SNPs at each frequency. Allele minor frequency distribution was transformed with a log scale because data were not normally distributed. The data were separated into two groups: synonymous and non-synonymous variants.
This graph shows the distribution of the two types of variants that interested us (synonymous and non-synonymous mutations) according to their allelic frequencies. As we can see, for a low minor frequency both variants seem to be more frequent and the missense variants tend to be higher than the synonymous one.
For a high minor allele frequency, SNPs seem to be less frequent and synonymous variants tend to be higher than the missense.
Using this plot, we were wondering if there was a difference in the SNP frequency spectrum for synonymous variants and for missense variants. Data representation was done with a stacked plot showing the proportion of the minor allele frequency for each type of variant.
The results showed a significant difference (p-value < 2.2e-16) with a Fisher-test comparing the two extremes bin.
The results were the following, SNPs at high frequency tend to have more synonymous mutations compared to low SNPs frequencies where they tend to be missense. This suggests that the mutations which do not change the amino acid (synonymous mutations) are less under a negative selective pressure than missense mutations. Therefore, they can spread more easily into the population and tend to be present at a higher proportion for high SNPs frequency. It is nevertheless necessary to remain attentive: we cannot talk about positive selection because as we said before we used minor allele frequency. Most likely SNPs at high frequency are more neutral and SNPs at low frequency are maybe more under the negative selection.
As we had the mutated position of the codons, we were interested in the distribution of the SNPs according to their frequencies by separating the groups with this new variable. To do this, we extracted the minor allele frequencies separated by the position of the codon change. At a high allele frequency, there is a bigger proportion of synonymous mutations. The type of mutation mostly depends on the mutated position of the codon. Therefore, we wanted to see if our previous result was consistent with the position of the codons that are mutated.
Here we plotted the SNPs frequencies on the x-axis and the percentage of the positions mutated within the codons on the y axis. We can see that the more a SNP is frequent in the population, the more it tends to have a mutation in the third position of the codon. This was expected because this is consistent with the result (part I SNPs), where we saw that at high SNP frequency, the proportion of synonymous mutations is higher. There are fewer chances to change the amino acid if the third position of the codon is mutated. On the contrary, if the first or the second position of a codon is mutated, it has more chances to change the amino acid encoded. To confirm it, we did a Fisher test for the 2 extreme bins (very low SNP frequency VS very high SNP frequency). The results show that the differences between the bins, that we see visually, are also statistically significant. The proportion of mutations at the third position is larger among the SNPs with higher frequencies. From this observation, we can speculate that mutations at the third position mostly induce synonymous mutations, due to the wobbling position. Therefore, they are more neutral which may imply that they are less under negative selection and allow them to spread into the population.
Part II: genes
In this part, we wondered if there were genes that have a higher percentage of synonymous mutations than others. We made the hypothesis that the genes with very specifics or critical functions would have more synonymous mutations as they are supposed to be under a strong purifying selection. We first plotted the frequencies of synonymous mutations for each gene, which provided us the following graph.
The difference between the black bars and one are the frequencies of missense mutations. On the extreme left, we have a group of genes that have 100% of synonymous mutations (383 genes). On the contrary, on the extreme right, there are genes with 0% of synonymous mutations, which means they have 100% of missense mutations (563 genes). We extracted the list of the genes with 100% synonymous mutations to have a look at their biological functions. Panterdb was used to submit the list of the gene that have 100% synonymous mutations (383 genes). We did a gene list analysis comparing to the reference genome. PANTHER GO slim biological process was used for the analysis to see if the genes we submitted were implied in important biological functions. The database allows us to display a graph.
The red bars are the percentage of genes involved in each function over the total number of genes in the reference list. In blue, this is the number of genes involved in each function over the total of genes we submitted.
As you can see, for each function indicated in the list on the left, there is a higher percentage for the genes we submitted than for the genes of the reference list. This suggests that the genes we submitted are not randomly involved in these functions. However, we need to be careful. These results showed that there is a correlation between the function of the gene and the frequency of synonymous mutations, but this is not causation. In other words, the function of the gene does not allow to infer about the frequency of synonymous mutations it may have.
Part III: tRNA relative abundances
In this part, we wanted to compare the relative abundance of tRNA in function of the codons. As we can observe in this plot, some codons have more tRNA available than others. tRNA relative abundances are not similar regarding the codon they recognize; this is called the codon bias. That means that there are optimal codons (codons with a higher tRNA gene copy number). We suggest that the selection may act on codon bias. This plot represents on the x-axis the 64 codons and on the y-axis the relative abundance of tRNA that can recognize them. The relative abundance of tRNAs that can recognize each codon is not uniform. Some codons have more tRNAs than others, even between codons coding for the same amino acid.
There are only 54 genes that encode tRNA anticodons in the human genome. The 10 remaining codons without complementary tRNA anticodon can be recognized due to the wobble position and the edited tRNA anticodon. To continue, we extracted the tRNA relative abundance associated with each codon (for the original and the mutated). Then, we did the absolute difference between the tRNA abundance for the original codon and for the mutated one. The difference gave us the delta abundance and we compared it with the associated frequency of the SNP.
5 plots were used to show the data separation. The first plot showed the median, the second the minimum to the first quantile, the third the first quantile to the median, the fourth the median to the third quantile, and finally the third quantile to the maximum.
Thanks to the data separation we were able to do a stacked plot for each separation in function of the minor allele frequency. The same colors used in the density plots were used.
We can observe that there is no pattern regarding the SNP frequency. With this plot we expected the results to show that the tRNA abundance does not depend on the SNP frequency.
For the statistical part, we did a Fisher test for the 2 extreme bins (very low SNP frequency VS very high SNP frequency).
We got two not significant (black) and two significant results (red). So not everything depends on the SNP frequencies. Even though we have two significant results, we could ask ourselves are those two really significant. Obviously, the results are not homogenous and do not correspond to what we expected.
References
(1) Cheon, M., Chang, I. and Hall, C.K. (2010), Extending the PRIME model for protein aggregation to all 20 amino acids. Proteins, 78: 2950-2960.
Contacts
melissa.monn@unil.ch
alicia.fauquex@unil.ch
aurelien.pirat@unil.ch