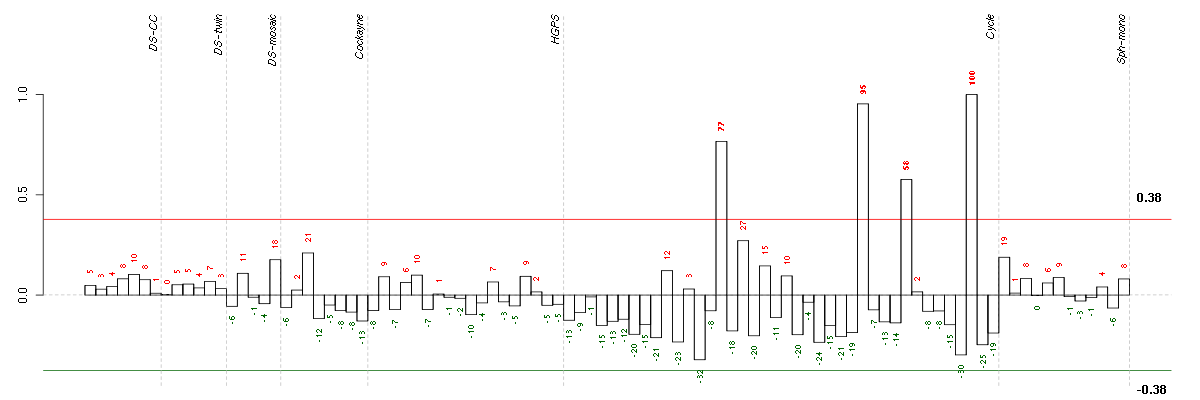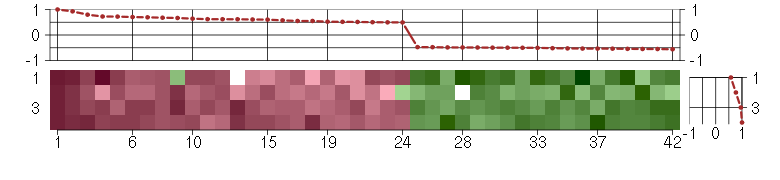

Under-expression is coded with green,
over-expression with red color.



AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: 0.55 AHI1Abelson helper integration site 1 (221569_at), score: -0.48 ANXA10annexin A10 (210143_at), score: -0.54 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.72 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.8 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.67 CAPZA1capping protein (actin filament) muscle Z-line, alpha 1 (208374_s_at), score: 0.6 CCNT2cyclin T2 (204645_at), score: -0.5 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.61 CHPcalcium binding protein P22 (214665_s_at), score: 0.62 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.55 CTTNcortactin (201059_at), score: 0.62 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.49 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.5 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: -0.53 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.55 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.73 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.49 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.5 IL33interleukin 33 (209821_at), score: -0.56 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: 0.52 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.5 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.7 NEFLneurofilament, light polypeptide (221805_at), score: -0.5 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.54 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.51 PARGpoly (ADP-ribose) glycohydrolase (205060_at), score: -0.5 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.53 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.57 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.48 RPL14ribosomal protein L14 (219138_at), score: 0.69 RPL27Aribosomal protein L27a (212044_s_at), score: 0.93 RPL38ribosomal protein L38 (202028_s_at), score: 0.68 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.64 RPS11ribosomal protein S11 (213350_at), score: 1 RPS19ribosomal protein S19 (202648_at), score: 0.55 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.52 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: -0.53 SLC35F5solute carrier family 35, member F5 (220123_at), score: -0.49 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: -0.49 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.62
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |