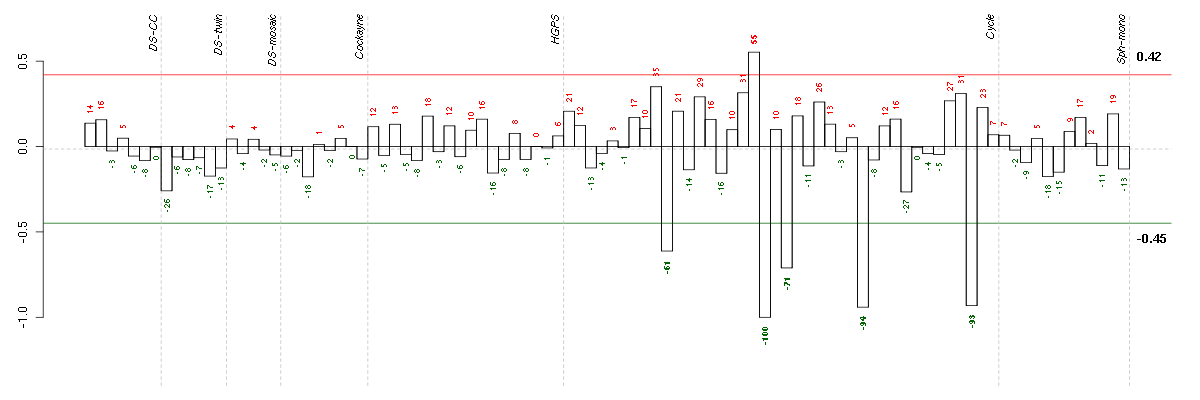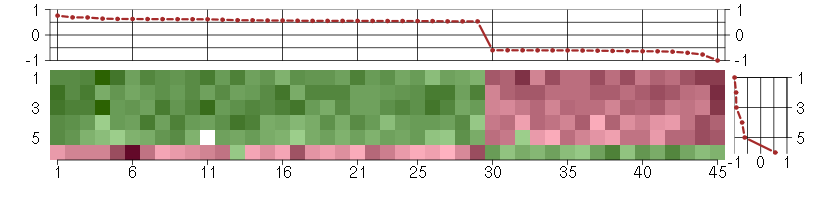

Under-expression is coded with green,
over-expression with red color.



ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.56 ANXA10annexin A10 (210143_at), score: 0.57 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: -0.6 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: -0.6 CD320CD320 molecule (218529_at), score: -0.64 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.55 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.63 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.69 CLUclusterin (208791_at), score: 0.55 COL5A3collagen, type V, alpha 3 (52255_s_at), score: 0.69 CST6cystatin E/M (206595_at), score: 0.58 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.62 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.55 DUSP6dual specificity phosphatase 6 (208891_at), score: 0.6 EDNRBendothelin receptor type B (204271_s_at), score: 0.53 EMCNendomucin (219436_s_at), score: -0.65 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.57 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.6 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.6 GPM6Bglycoprotein M6B (209167_at), score: 0.62 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.7 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.54 HSPB2heat shock 27kDa protein 2 (205824_at), score: -0.6 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.55 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: 0.58 IL33interleukin 33 (209821_at), score: 0.76 KRT33Akeratin 33A (208483_x_at), score: 0.55 LOC388152hypothetical LOC388152 (220602_s_at), score: -0.62 MLPHmelanophilin (218211_s_at), score: -0.64 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: 0.53 NEFLneurofilament, light polypeptide (221805_at), score: 0.62 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.54 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: 0.55 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: 0.64 PRSS3protease, serine, 3 (213421_x_at), score: 0.62 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.55 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: 0.55 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: -0.62 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.61 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.77 SYT11synaptotagmin XI (209198_s_at), score: -0.6 TMSB15Athymosin beta 15a (205347_s_at), score: -0.64 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.62 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -1 ZNF219zinc finger protein 219 (219314_s_at), score: 0.63
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |