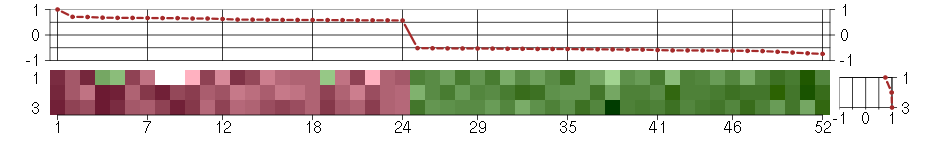

Under-expression is coded with green,
over-expression with red color.



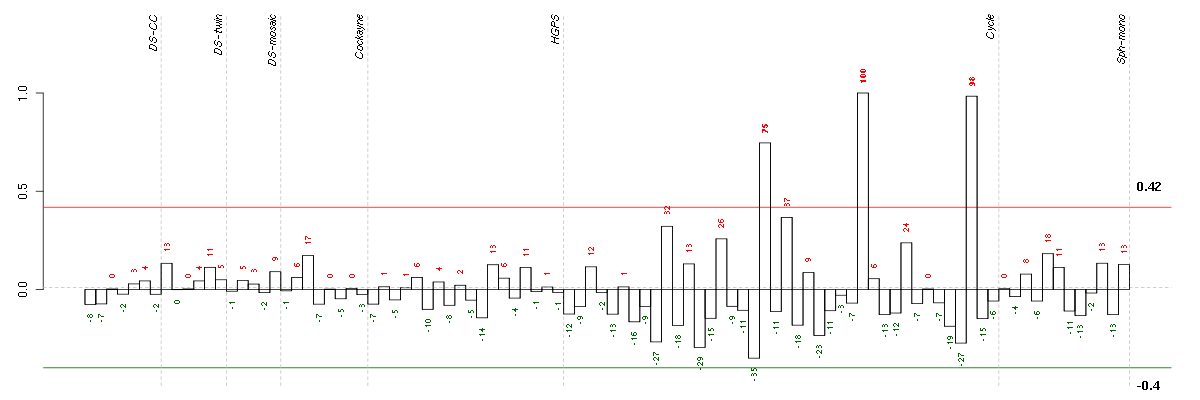
ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.54 ANXA10annexin A10 (210143_at), score: -0.57 ARL17ADP-ribosylation factor-like 17 (210435_at), score: -0.57 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.71 BAXBCL2-associated X protein (208478_s_at), score: 0.57 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.59 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.66 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.66 CD320CD320 molecule (218529_at), score: 0.71 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.53 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.61 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.63 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.69 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.59 CST6cystatin E/M (206595_at), score: -0.53 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.61 EMCNendomucin (219436_s_at), score: 0.59 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.62 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.58 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.54 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.65 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.6 GANgigaxonin (220124_at), score: -0.56 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.67 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.6 GPSM3G-protein signaling modulator 3 (AGS3-like, C. elegans) (204265_s_at), score: 0.58 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.6 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.57 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.52 IL33interleukin 33 (209821_at), score: -0.74 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.55 MLPHmelanophilin (218211_s_at), score: 0.58 NEFLneurofilament, light polypeptide (221805_at), score: -0.62 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.61 NMBneuromedin B (205204_at), score: 0.59 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.63 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.53 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: -0.53 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.56 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.72 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.55 RPL14ribosomal protein L14 (219138_at), score: 0.59 RPL27Aribosomal protein L27a (212044_s_at), score: 0.67 RPS11ribosomal protein S11 (213350_at), score: 0.68 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.57 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.67 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.6 TMSB15Athymosin beta 15a (205347_s_at), score: 0.65 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.66 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 ZNF219zinc finger protein 219 (219314_s_at), score: -0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |