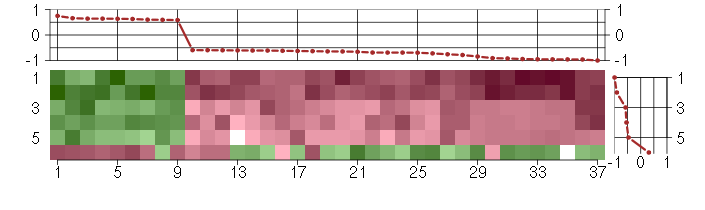

Under-expression is coded with green,
over-expression with red color.



A2Malpha-2-macroglobulin (217757_at), score: -0.65 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -1 AIM2absent in melanoma 2 (206513_at), score: -0.64 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.59 CD24CD24 molecule (266_s_at), score: -0.61 CRTAPcartilage associated protein (201380_at), score: 0.64 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.76 DIO2deiodinase, iodothyronine, type II (203700_s_at), score: -0.59 EPHB2EPH receptor B2 (209589_s_at), score: -0.6 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.62 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.75 GAGE4G antigen 4 (207086_x_at), score: -0.96 GAGE6G antigen 6 (208155_x_at), score: -0.91 GATA3GATA binding protein 3 (209604_s_at), score: -0.84 HERC5hect domain and RLD 5 (219863_at), score: -0.69 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.64 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.6 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.79 MYO1Cmyosin IC (214656_x_at), score: 0.6 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.63 NEFLneurofilament, light polypeptide (221805_at), score: -0.63 NMUneuromedin U (206023_at), score: -0.61 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.69 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.96 PDE10Aphosphodiesterase 10A (205501_at), score: -0.66 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.61 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.64 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 0.66 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.69 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.69 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.92 TMOD1tropomodulin 1 (203661_s_at), score: -0.6 UBL3ubiquitin-like 3 (201535_at), score: 0.63 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.94 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.96 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.95 WT1Wilms tumor 1 (206067_s_at), score: -0.72
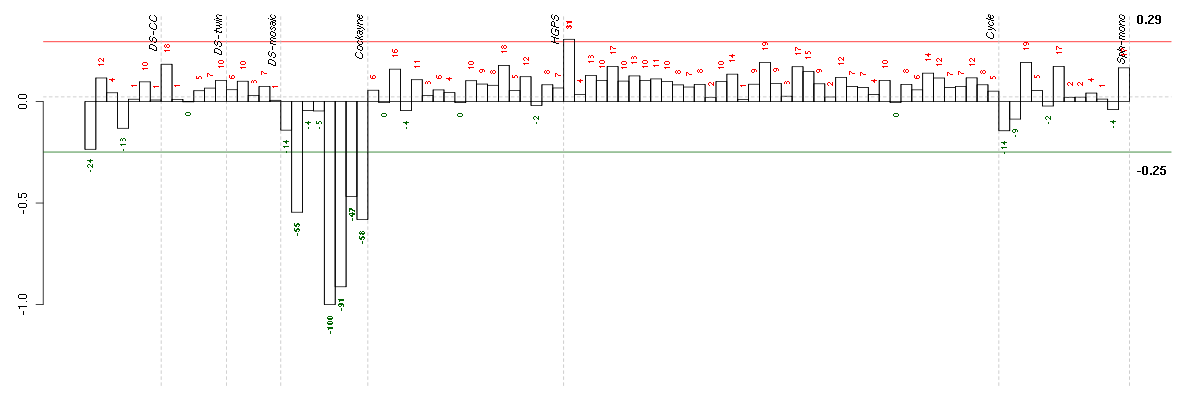
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |