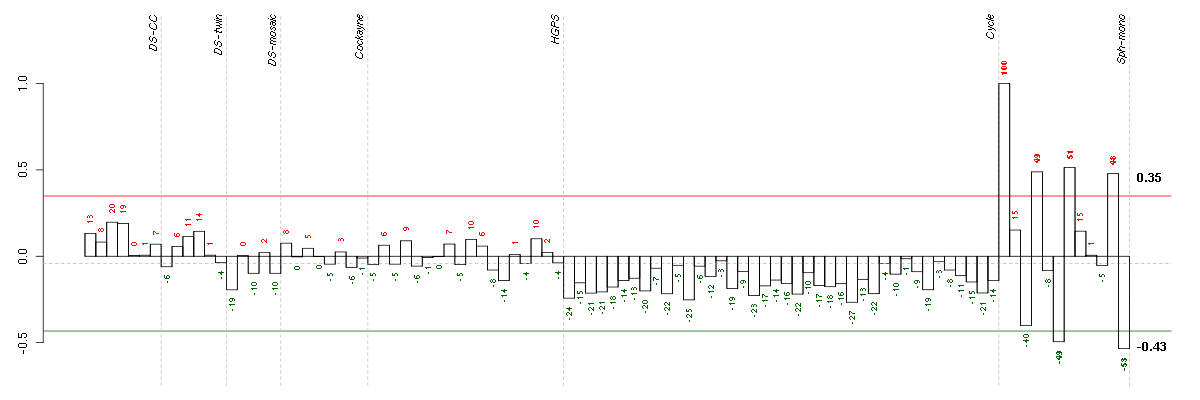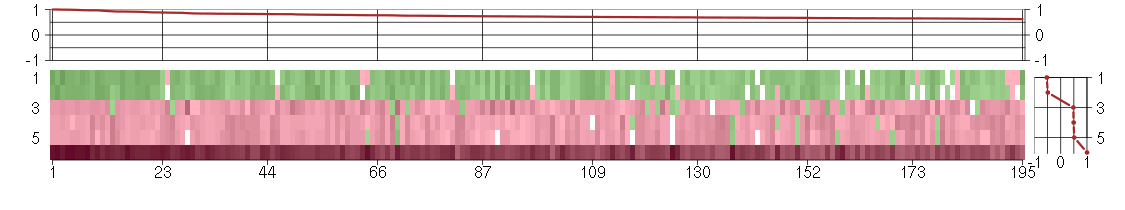

Under-expression is coded with green,
over-expression with red color.



ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.75 ACOT11acyl-CoA thioesterase 11 (216103_at), score: 0.73 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.86 AKAP8A kinase (PRKA) anchor protein 8 (203848_at), score: 0.65 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: 0.78 ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.92 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: 0.73 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.97 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (202548_s_at), score: 0.77 BCL9B-cell CLL/lymphoma 9 (204129_at), score: 0.81 BCORBCL6 co-repressor (219433_at), score: 0.63 BTF3L2basic transcription factor 3, like 2 (217461_x_at), score: 0.68 C10orf88chromosome 10 open reading frame 88 (219240_s_at), score: 0.63 C11orf57chromosome 11 open reading frame 57 (218314_s_at), score: 0.8 C12orf35chromosome 12 open reading frame 35 (218614_at), score: 0.63 C12orf49chromosome 12 open reading frame 49 (218867_s_at), score: 0.65 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.93 C6orf106chromosome 6 open reading frame 106 (217925_s_at), score: 0.68 C6orf162chromosome 6 open reading frame 162 (213314_at), score: 0.65 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: 0.82 CCDC25coiled-coil domain containing 25 (218125_s_at), score: 0.7 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (205288_at), score: 0.81 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: 0.95 CEP27centrosomal protein 27kDa (220071_x_at), score: 0.64 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: 0.78 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.76 CIRCBF1 interacting corepressor (209571_at), score: 0.67 CIRBPcold inducible RNA binding protein (200811_at), score: 0.72 CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: 0.83 CNOT6CCR4-NOT transcription complex, subunit 6 (217970_s_at), score: 0.7 CNOT8CCR4-NOT transcription complex, subunit 8 (202163_s_at), score: 0.69 CPEcarboxypeptidase E (201117_s_at), score: 0.73 CREBZFCREB/ATF bZIP transcription factor (202978_s_at), score: 0.69 CYP1A2cytochrome P450, family 1, subfamily A, polypeptide 2 (207608_x_at), score: 0.71 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: 0.72 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.92 DAZ4deleted in azoospermia 4 (216351_x_at), score: 0.71 DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (204909_at), score: 0.66 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.73 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: 0.74 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: 0.97 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: 0.73 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.87 FAM134Afamily with sequence similarity 134, member A (222129_at), score: 0.83 FAM53Bfamily with sequence similarity 53, member B (203206_at), score: 0.67 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.74 FBXO41F-box protein 41 (44040_at), score: 0.74 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: 0.84 FCARFc fragment of IgA, receptor for (211305_x_at), score: 0.83 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: 0.7 FKSG49FKSG49 (211454_x_at), score: 0.71 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 1 FLJ23172hypothetical LOC389177 (217016_x_at), score: 0.8 FLJ42627hypothetical LOC645644 (220352_x_at), score: 0.71 FNBP4formin binding protein 4 (212232_at), score: 0.83 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: 0.73 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.99 GCLCglutamate-cysteine ligase, catalytic subunit (202922_at), score: 0.74 GIGYF2GRB10 interacting GYF protein 2 (212260_at), score: 0.68 GPATCH8G patch domain containing 8 (212485_at), score: 0.7 GTPBP3GTP binding protein 3 (mitochondrial) (213835_x_at), score: 0.74 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: 0.91 HECAheadcase homolog (Drosophila) (218603_at), score: 0.66 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: 0.84 HIC2hypermethylated in cancer 2 (212964_at), score: 0.66 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: 0.78 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (203285_s_at), score: 0.67 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.87 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.65 IMAASLC7A5 pseudogene (208118_x_at), score: 0.74 INSRinsulin receptor (213792_s_at), score: 0.72 IREB2iron-responsive element binding protein 2 (214666_x_at), score: 0.65 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.83 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.68 KIAA0894KIAA0894 protein (207436_x_at), score: 0.66 KIF1Bkinesin family member 1B (209234_at), score: 0.65 KLF11Kruppel-like factor 11 (218486_at), score: 0.66 KLHL24kelch-like 24 (Drosophila) (206551_x_at), score: 0.65 LETM1leucine zipper-EF-hand containing transmembrane protein 1 (222006_at), score: 0.66 LIMK1LIM domain kinase 1 (204357_s_at), score: 0.68 LOC100128701heterogeneous nuclear ribonucleoprotein A1-like 2 pseudogene (216497_at), score: 0.7 LOC100128836similar to heterogeneous nuclear ribonucleoprotein A1 (217353_at), score: 0.67 LOC100129624hypothetical LOC100129624 (210748_at), score: 0.69 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: 0.78 LOC152719hypothetical protein LOC152719 (215978_x_at), score: 0.72 LOC171220destrin-2 pseudogene (211325_x_at), score: 0.66 LOC51152melanoma antigen (220771_at), score: 0.65 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: 0.92 LOC644617hypothetical LOC644617 (221235_s_at), score: 0.82 LOC645139hypothetical LOC645139 (209064_x_at), score: 0.69 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.9 LOC728844hypothetical LOC728844 (222040_at), score: 0.64 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: 0.69 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: 0.84 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.64 MCL1myeloid cell leukemia sequence 1 (BCL2-related) (214057_at), score: 0.68 MDN1MDN1, midasin homolog (yeast) (212693_at), score: 0.63 MEFVMediterranean fever (208262_x_at), score: 0.76 METAP2methionyl aminopeptidase 2 (202015_x_at), score: 0.84 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.89 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.75 MLLT10myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 (216506_x_at), score: 0.76 MRPL44mitochondrial ribosomal protein L44 (218202_x_at), score: 0.78 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.73 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.64 OPHN1oligophrenin 1 (206323_x_at), score: 0.73 PARP6poly (ADP-ribose) polymerase family, member 6 (219639_x_at), score: 0.68 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.82 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: 0.7 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.79 PER2period homolog 2 (Drosophila) (205251_at), score: 0.71 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.65 PGFplacental growth factor (215179_x_at), score: 0.7 PIGAphosphatidylinositol glycan anchor biosynthesis, class A (205281_s_at), score: 0.67 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: 0.75 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: 0.72 PODNL1podocan-like 1 (220411_x_at), score: 0.67 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.82 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.83 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.73 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.77 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: 0.82 RAD52RAD52 homolog (S. cerevisiae) (205647_at), score: 0.65 RGL1ral guanine nucleotide dissociation stimulator-like 1 (209568_s_at), score: 0.71 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.71 RHEBRas homolog enriched in brain (213409_s_at), score: 0.72 RNMTRNA (guanine-7-) methyltransferase (202683_s_at), score: 0.79 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: 0.85 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.66 RPL35Aribosomal protein L35a (215208_x_at), score: 0.79 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: 0.69 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (214764_at), score: 0.68 SAP30LSAP30-like (219129_s_at), score: 0.66 SAPS2SAPS domain family, member 2 (202792_s_at), score: 0.76 SAPS3SAPS domain family, member 3 (217928_s_at), score: 0.68 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.63 SCD5stearoyl-CoA desaturase 5 (220232_at), score: 0.8 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: 0.68 SETD4SET domain containing 4 (219482_at), score: 0.64 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: 0.79 SFRS18splicing factor, arginine/serine-rich 18 (212176_at), score: 0.71 SFTPBsurfactant protein B (214354_x_at), score: 0.66 SH3BP2SH3-domain binding protein 2 (217257_at), score: 0.88 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: 0.97 SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.92 SLC11A1solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 (217473_x_at), score: 0.73 SLC25A44solute carrier family 25, member 44 (32091_at), score: 0.71 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.98 SLC35F2solute carrier family 35, member F2 (218826_at), score: 0.7 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: 0.69 SMEK2SMEK homolog 2, suppressor of mek1 (Dictyostelium) (222270_at), score: 0.67 SNX15sorting nexin 15 (205482_x_at), score: 0.68 SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.83 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.75 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.94 SPNsialophorin (206057_x_at), score: 0.99 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 0.99 STX3syntaxin 3 (209238_at), score: 0.63 SYCP1synaptonemal complex protein 1 (206740_x_at), score: 0.68 TBC1D15TBC1 domain family, member 15 (218268_at), score: 0.66 TCTN2tectonic family member 2 (206438_x_at), score: 0.82 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 1 TJAP1tight junction associated protein 1 (peripheral) (47608_at), score: 0.64 TMEM38Btransmembrane protein 38B (218772_x_at), score: 0.65 TP53TG3TP53 target 3 (220167_s_at), score: 0.65 TRIM36tripartite motif-containing 36 (219736_at), score: 0.76 TRIT1tRNA isopentenyltransferase 1 (218617_at), score: 0.71 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.63 UBAP1ubiquitin associated protein 1 (221490_at), score: 0.66 UBQLN4ubiquilin 4 (222252_x_at), score: 0.87 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: 0.65 UPP1uridine phosphorylase 1 (203234_at), score: 0.77 USP12ubiquitin specific peptidase 12 (213327_s_at), score: 0.69 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.97 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: 0.67 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.79 YTHDF1YTH domain family, member 1 (221741_s_at), score: 0.68 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.88 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.91 ZNF160zinc finger protein 160 (214715_x_at), score: 0.7 ZNF253zinc finger protein 253 (206900_x_at), score: 0.62 ZNF432zinc finger protein 432 (219848_s_at), score: 0.7 ZNF440zinc finger protein 440 (215892_at), score: 0.65 ZNF451zinc finger protein 451 (212557_at), score: 0.67 ZNF468zinc finger protein 468 (214751_at), score: 0.64 ZNF492zinc finger protein 492 (215532_x_at), score: 0.74 ZNF506zinc finger protein 506 (221626_at), score: 0.7 ZNF528zinc finger protein 528 (215019_x_at), score: 0.82 ZNF552zinc finger protein 552 (219741_x_at), score: 0.76 ZNF611zinc finger protein 611 (208137_x_at), score: 0.77 ZNF639zinc finger protein 639 (218413_s_at), score: 0.83 ZNF770zinc finger protein 770 (220608_s_at), score: 0.7 ZNF816Azinc finger protein 816A (217541_x_at), score: 0.89 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |