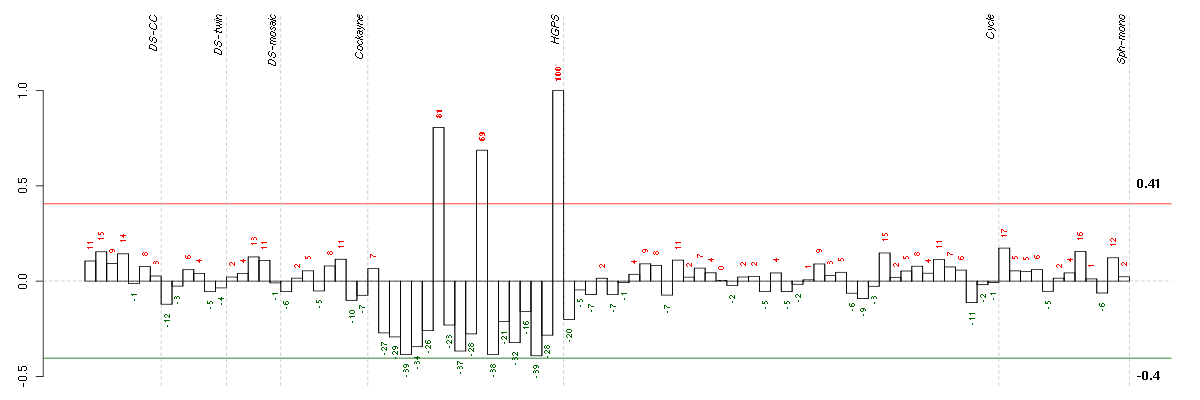

Under-expression is coded with green,
over-expression with red color.

defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.71 ABCC4ATP-binding cassette, sub-family C (CFTR/MRP), member 4 (203196_at), score: 0.51 ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: 0.52 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.5 ADAMTSL3ADAMTS-like 3 (213974_at), score: 0.73 ADMadrenomedullin (202912_at), score: -0.65 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.71 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: -0.61 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: -0.54 AMFRautocrine motility factor receptor (202203_s_at), score: 0.55 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.72 ANO3anoctamin 3 (215241_at), score: -0.56 ANXA10annexin A10 (210143_at), score: 0.61 ANXA3annexin A3 (209369_at), score: 0.71 ARSBarylsulfatase B (206129_s_at), score: 0.56 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: -0.59 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (203304_at), score: -0.54 BMP6bone morphogenetic protein 6 (206176_at), score: 0.55 BNC2basonuclin 2 (220272_at), score: 0.69 C3complement component 3 (217767_at), score: 0.56 C8orf84chromosome 8 open reading frame 84 (214725_at), score: 0.82 CADPS2Ca++-dependent secretion activator 2 (219572_at), score: 0.6 CCDC68coiled-coil domain containing 68 (220180_at), score: 0.56 CD200CD200 molecule (209583_s_at), score: 0.85 CD34CD34 molecule (209543_s_at), score: 0.74 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.62 CD70CD70 molecule (206508_at), score: 0.58 CEBPGCCAAT/enhancer binding protein (C/EBP), gamma (204203_at), score: 0.51 CENPQcentromere protein Q (219294_at), score: 0.49 CFHcomplement factor H (213800_at), score: 0.8 CFHR1complement factor H-related 1 (215388_s_at), score: 0.67 CHRDL1chordin-like 1 (209763_at), score: 0.67 CLDN1claudin 1 (218182_s_at), score: 0.68 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.68 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.5 CRYBB2crystallin, beta B2 (206777_s_at), score: 0.59 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.65 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.76 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.62 DAAM1dishevelled associated activator of morphogenesis 1 (216060_s_at), score: 0.63 DDX43DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 (220004_at), score: 0.63 DLEU1deleted in lymphocytic leukemia 1 (non-protein coding) (205677_s_at), score: 0.6 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.55 DOCK9dedicator of cytokinesis 9 (212538_at), score: 0.54 DPYSL3dihydropyrimidinase-like 3 (201431_s_at), score: 0.55 DSG2desmoglein 2 (217901_at), score: 0.75 DUSP4dual specificity phosphatase 4 (204014_at), score: 0.55 EDN1endothelin 1 (218995_s_at), score: 0.64 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.57 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.57 EN1engrailed homeobox 1 (220559_at), score: -0.71 EPHA5EPH receptor A5 (215664_s_at), score: 0.75 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: 0.54 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (213541_s_at), score: 0.84 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.56 FBLN2fibulin 2 (203886_s_at), score: -0.7 FGL2fibrinogen-like 2 (204834_at), score: 1 FKBP1BFK506 binding protein 1B, 12.6 kDa (206857_s_at), score: 0.49 FLI1Friend leukemia virus integration 1 (204236_at), score: 0.51 FMO3flavin containing monooxygenase 3 (40665_at), score: 0.67 FOXD1forkhead box D1 (206307_s_at), score: -0.55 FOXO1forkhead box O1 (202724_s_at), score: 0.57 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.65 GPR1G protein-coupled receptor 1 (214605_x_at), score: 0.51 GPR126G protein-coupled receptor 126 (213094_at), score: 0.77 GPR4G protein-coupled receptor 4 (206236_at), score: 0.66 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.68 GSTT2glutathione S-transferase theta 2 (205439_at), score: 0.73 HDAC5histone deacetylase 5 (202455_at), score: -0.54 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.59 HRH1histamine receptor H1 (205580_s_at), score: 0.59 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: -0.7 HYIhydroxypyruvate isomerase homolog (E. coli) (221435_x_at), score: 0.67 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.74 IL12Ainterleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) (207160_at), score: -0.55 IL18interleukin 18 (interferon-gamma-inducing factor) (206295_at), score: 0.73 IL1Ainterleukin 1, alpha (210118_s_at), score: 0.7 IL1Binterleukin 1, beta (39402_at), score: 0.54 IL21Rinterleukin 21 receptor (219971_at), score: 0.51 IL8interleukin 8 (211506_s_at), score: 0.55 ITM2Aintegral membrane protein 2A (202746_at), score: 0.72 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.52 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: 0.63 KIAA0182KIAA0182 (212057_at), score: -0.56 KIAA0247KIAA0247 (202181_at), score: 0.51 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: 0.59 KRT8keratin 8 (209008_x_at), score: 0.52 LAMC2laminin, gamma 2 (202267_at), score: 0.81 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: -0.56 LIPGlipase, endothelial (219181_at), score: 0.9 LOC100132214similar to HSPC047 protein (220692_at), score: 0.51 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: 0.74 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: -0.58 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: 0.66 MFAP2microfibrillar-associated protein 2 (203417_at), score: -0.68 MFGE8milk fat globule-EGF factor 8 protein (210605_s_at), score: -0.55 MYLKmyosin light chain kinase (202555_s_at), score: -0.73 NAAAN-acylethanolamine acid amidase (214765_s_at), score: -0.57 NFIBnuclear factor I/B (209290_s_at), score: 0.66 NOX4NADPH oxidase 4 (219773_at), score: 0.79 OSBPL1Aoxysterol binding protein-like 1A (209485_s_at), score: -0.62 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: 0.49 PARP3poly (ADP-ribose) polymerase family, member 3 (209940_at), score: -0.7 PDE10Aphosphodiesterase 10A (205501_at), score: 0.53 PDE4Dphosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) (204491_at), score: 0.52 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.62 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: -0.55 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: 0.67 PKP2plakophilin 2 (207717_s_at), score: 0.64 PLCB4phospholipase C, beta 4 (203896_s_at), score: -0.59 PLCL1phospholipase C-like 1 (205934_at), score: 0.51 PLXNA2plexin A2 (213030_s_at), score: 0.76 POPDC3popeye domain containing 3 (219926_at), score: -0.7 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: -0.65 PPM1Hprotein phosphatase 1H (PP2C domain containing) (212686_at), score: 0.68 PSPHphosphoserine phosphatase (205048_s_at), score: 0.55 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.49 RAC2ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) (213603_s_at), score: -0.67 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: 0.56 RPP25ribonuclease P/MRP 25kDa subunit (219143_s_at), score: -0.58 RXRAretinoid X receptor, alpha (202449_s_at), score: -0.55 SCRN1secernin 1 (201462_at), score: -0.69 SLC17A9solute carrier family 17, member 9 (219559_at), score: -0.62 SLC31A2solute carrier family 31 (copper transporters), member 2 (204204_at), score: 0.58 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: 0.73 SLC7A5solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 (201195_s_at), score: 0.52 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: 0.84 STBD1starch binding domain 1 (203986_at), score: -0.64 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.6 SUSD5sushi domain containing 5 (214954_at), score: 0.55 SWAP70SWAP-70 protein (209307_at), score: 0.53 SYCP2synaptonemal complex protein 2 (206546_at), score: -0.6 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.5 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.49 THBS2thrombospondin 2 (203083_at), score: 0.54 THY1Thy-1 cell surface antigen (208850_s_at), score: -0.71 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: -0.7 TLR4toll-like receptor 4 (221060_s_at), score: 0.81 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.63 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.69 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.71 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (214755_at), score: -0.58 VLDLRvery low density lipoprotein receptor (209822_s_at), score: 0.6 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.55 WDTC1WD and tetratricopeptide repeats 1 (40829_at), score: -0.57 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.5 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.62 ZFHX4zinc finger homeobox 4 (219779_at), score: 0.75 ZFPM2zinc finger protein, multitype 2 (219778_at), score: 0.5 ZNF323zinc finger protein 323 (222016_s_at), score: 0.71 ZNF365zinc finger protein 365 (206448_at), score: 0.62
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |