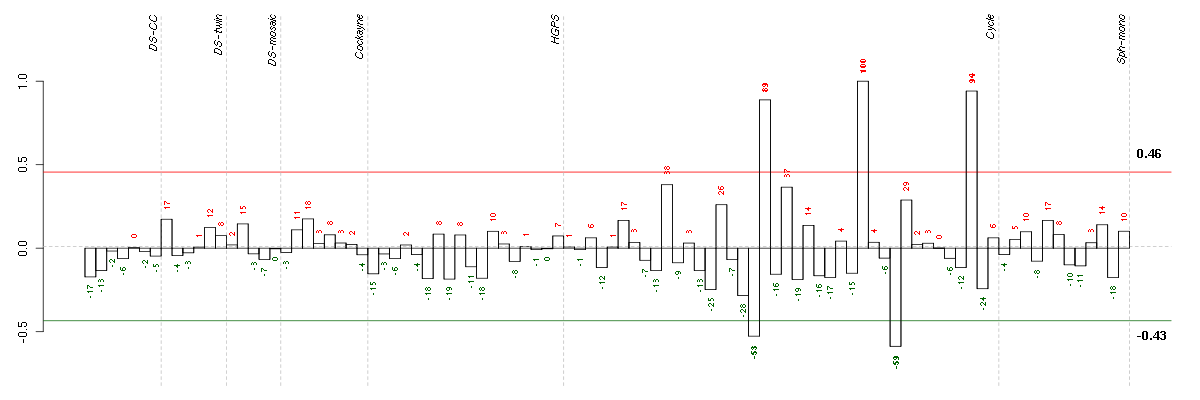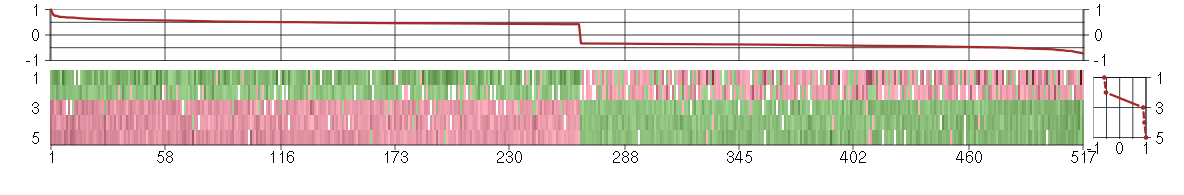

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (212772_s_at), score: -0.42 ACANaggrecan (205679_x_at), score: 0.59 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.55 ADAMTSL4ADAMTS-like 4 (220578_at), score: -0.36 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.46 ADSSadenylosuccinate synthase (221761_at), score: -0.36 AFF2AF4/FMR2 family, member 2 (206105_at), score: -0.55 AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: 0.43 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.61 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.36 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.46 ANKHD1ankyrin repeat and KH domain containing 1 (208773_s_at), score: 0.42 ANKRD28ankyrin repeat domain 28 (213035_at), score: 0.53 ANKRD55ankyrin repeat domain 55 (220112_at), score: -0.42 ANXA10annexin A10 (210143_at), score: -0.6 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: 0.56 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.43 ARL17ADP-ribosylation factor-like 17 (210435_at), score: -0.41 ARMC9armadillo repeat containing 9 (219637_at), score: 0.51 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.64 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.44 ASCC1activating signal cointegrator 1 complex subunit 1 (219336_s_at), score: 0.42 ASTN2astrotactin 2 (209693_at), score: 0.46 ATG2BATG2 autophagy related 2 homolog B (S. cerevisiae) (219164_s_at), score: 0.45 ATN1atrophin 1 (40489_at), score: -0.43 ATRNL1attractin-like 1 (213744_at), score: -0.43 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: -0.34 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.71 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.46 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: -0.39 BAXBCL2-associated X protein (208478_s_at), score: 0.58 BCAT2branched chain aminotransferase 2, mitochondrial (203576_at), score: 0.43 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.57 BCL11AB-cell CLL/lymphoma 11A (zinc finger protein) (219497_s_at), score: -0.41 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (219528_s_at), score: -0.46 BCS1LBCS1-like (yeast) (207618_s_at), score: 0.48 BIN3bridging integrator 3 (222199_s_at), score: 0.46 BMP1bone morphogenetic protein 1 (207595_s_at), score: -0.36 BMP7bone morphogenetic protein 7 (209590_at), score: -0.43 BTBD3BTB (POZ) domain containing 3 (202946_s_at), score: -0.37 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.45 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.5 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: 0.44 C11orf51chromosome 11 open reading frame 51 (204218_at), score: 0.42 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.58 C12orf52chromosome 12 open reading frame 52 (221777_at), score: 0.45 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.37 C16orf62chromosome 16 open reading frame 62 (203173_s_at), score: 0.44 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.57 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.41 C19orf54chromosome 19 open reading frame 54 (222052_at), score: 0.52 C1orf159chromosome 1 open reading frame 159 (219337_at), score: -0.42 C1orf50chromosome 1 open reading frame 50 (219406_at), score: 0.46 C1orf69chromosome 1 open reading frame 69 (215490_at), score: 0.42 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.47 C2CD2LC2CD2-like (204757_s_at), score: -0.55 C2orf49chromosome 2 open reading frame 49 (219662_at), score: 0.45 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.34 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.68 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.44 C8orf41chromosome 8 open reading frame 41 (219124_at), score: 0.54 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.68 C9orf61chromosome 9 open reading frame 61 (213900_at), score: -0.44 CA1carbonic anhydrase I (205949_at), score: -0.54 CA12carbonic anhydrase XII (203963_at), score: 0.52 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.34 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.52 CAPN9calpain 9 (210641_at), score: 0.45 CAPZA1capping protein (actin filament) muscle Z-line, alpha 1 (208374_s_at), score: 0.6 CCDC88Acoiled-coil domain containing 88A (219387_at), score: 0.43 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.58 CCNE1cyclin E1 (213523_at), score: -0.34 CCNT2cyclin T2 (204645_at), score: -0.34 CD226CD226 molecule (207315_at), score: -0.42 CD320CD320 molecule (218529_at), score: 0.68 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.47 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: -0.42 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.39 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.43 CDCA4cell division cycle associated 4 (218399_s_at), score: -0.43 CDCP1CUB domain containing protein 1 (218451_at), score: -0.43 CDH18cadherin 18, type 2 (206280_at), score: 0.5 CDKAL1CDK5 regulatory subunit associated protein 1-like 1 (214877_at), score: 0.45 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: 0.51 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.34 CENPTcentromere protein T (218148_at), score: -0.43 CEP70centrosomal protein 70kDa (219036_at), score: 0.52 CHPcalcium binding protein P22 (214665_s_at), score: 0.48 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: 0.47 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.51 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.57 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.49 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.36 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: -0.47 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.61 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.39 CLPBClpB caseinolytic peptidase B homolog (E. coli) (221845_s_at), score: 0.46 CLUclusterin (208791_at), score: -0.44 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.42 CNNM2cyclin M2 (206818_s_at), score: 0.6 CNPY3canopy 3 homolog (zebrafish) (217931_at), score: 0.46 COASYCoenzyme A synthase (201913_s_at), score: 0.45 COG2component of oligomeric golgi complex 2 (203073_at), score: 0.48 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.38 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.68 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.43 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.48 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (213379_at), score: 0.46 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: 0.5 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.63 CR1complement component (3b/4b) receptor 1 (Knops blood group) (217484_at), score: 0.51 CROCCL1ciliary rootlet coiled-coil, rootletin-like 1 (211038_s_at), score: 0.46 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.36 CSPG4chondroitin sulfate proteoglycan 4 (214297_at), score: 0.43 CST6cystatin E/M (206595_at), score: -0.42 CTRLchymotrypsin-like (214377_s_at), score: -0.37 CTSKcathepsin K (202450_s_at), score: -0.38 CTSOcathepsin O (203758_at), score: -0.49 CTSScathepsin S (202901_x_at), score: -0.36 CTSZcathepsin Z (210042_s_at), score: 0.58 CTTNcortactin (201059_at), score: 0.51 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.45 CYLC2cylicin, basic protein of sperm head cytoskeleton 2 (207780_at), score: -0.4 D4S234EDNA segment on chromosome 4 (unique) 234 expressed sequence (209569_x_at), score: -0.43 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.5 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.48 DCPSdecapping enzyme, scavenger (218774_at), score: 0.47 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: 0.42 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: -0.37 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.59 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.48 DNAJC8DnaJ (Hsp40) homolog, subfamily C, member 8 (205545_x_at), score: 0.46 DNM3dynamin 3 (209839_at), score: -0.54 DOM3Zdom-3 homolog Z (C. elegans) (215982_s_at), score: 0.49 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.43 DSCR3Down syndrome critical region gene 3 (203635_at), score: 0.53 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.37 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.5 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.48 DUSP6dual specificity phosphatase 6 (208891_at), score: -0.54 DVL2dishevelled, dsh homolog 2 (Drosophila) (57532_at), score: 0.45 DXS542X-linked retinopathy protein-like (222247_at), score: -0.37 EDNRBendothelin receptor type B (204271_s_at), score: -0.43 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.36 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: -0.38 EGR3early growth response 3 (206115_at), score: -0.36 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: -0.34 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.46 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: -0.44 ELA3Aelastase 3A, pancreatic (211738_x_at), score: -0.45 ELNelastin (212670_at), score: -0.36 EMCNendomucin (219436_s_at), score: 0.66 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.46 EPOerythropoietin (217254_s_at), score: 0.55 EREGepiregulin (205767_at), score: -0.4 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.35 ESRRAestrogen-related receptor alpha (1487_at), score: -0.44 EVI2Aecotropic viral integration site 2A (204774_at), score: 0.47 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.61 EVLEnah/Vasp-like (217838_s_at), score: -0.38 EXOSC5exosome component 5 (218481_at), score: 0.64 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: 0.49 FAM131Bfamily with sequence similarity 131, member B (205368_at), score: -0.38 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.35 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.4 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: 0.43 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.55 FAM70Afamily with sequence similarity 70, member A (219895_at), score: -0.39 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.51 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.36 FBXO31F-box protein 31 (219785_s_at), score: 0.53 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: 0.49 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.39 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.48 FKSG49FKSG49 (211454_x_at), score: 0.45 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.5 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.43 FOXK2forkhead box K2 (203064_s_at), score: -0.39 FUZfuzzy homolog (Drosophila) (221187_s_at), score: 0.48 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.52 GAAglucosidase, alpha; acid (202812_at), score: -0.49 GAGE4G antigen 4 (207086_x_at), score: -0.33 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.51 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: 0.47 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.51 GANgigaxonin (220124_at), score: -0.53 GBAPglucosidase, beta; acid, pseudogene (210589_s_at), score: 0.45 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.44 GDF9growth differentiation factor 9 (221314_at), score: -0.46 GKglycerol kinase (207387_s_at), score: -0.37 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.66 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: 0.44 GPM6Bglycoprotein M6B (209167_at), score: -0.51 GPR153G protein-coupled receptor 153 (221902_at), score: -0.47 GPR172AG protein-coupled receptor 172A (222155_s_at), score: -0.39 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.43 GPR27G protein-coupled receptor 27 (221306_at), score: 0.5 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.68 GPSM3G-protein signaling modulator 3 (AGS3-like, C. elegans) (204265_s_at), score: 0.56 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: 0.5 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.44 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.35 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: 0.55 GTPBP6GTP binding protein 6 (putative) (203314_at), score: 0.44 H2AFJH2A histone family, member J (220936_s_at), score: -0.48 HEATR6HEAT repeat containing 6 (65493_at), score: 0.45 HERC3hect domain and RLD 3 (206183_s_at), score: -0.39 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.47 HINFPhistone H4 transcription factor (206495_s_at), score: 0.61 HISPPD2Ahistidine acid phosphatase domain containing 2A (204578_at), score: 0.57 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: 0.54 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.36 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: -0.42 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: -0.44 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: -0.33 HNRNPUL2heterogeneous nuclear ribonucleoprotein U-like 2 (66053_at), score: 0.46 HOXD1homeobox D1 (205975_s_at), score: -0.38 HSD11B1hydroxysteroid (11-beta) dehydrogenase 1 (205404_at), score: -0.41 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.59 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: 0.46 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.37 IER2immediate early response 2 (202081_at), score: -0.48 IFNA5interferon, alpha 5 (214569_at), score: -0.37 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.49 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.48 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.42 IL33interleukin 33 (209821_at), score: -0.73 INTS12integrator complex subunit 12 (218616_at), score: 0.42 INTS8integrator complex subunit 8 (218905_at), score: 0.52 IRF6interferon regulatory factor 6 (202597_at), score: -0.4 ISOC2isochorismatase domain containing 2 (218893_at), score: 0.43 JMJD1Ajumonji domain containing 1A (212689_s_at), score: 0.44 JUNBjun B proto-oncogene (201473_at), score: -0.42 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.33 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.56 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: -0.37 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.34 KCTD14potassium channel tetramerisation domain containing 14 (58916_at), score: 0.59 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.34 KIAA0415KIAA0415 (209912_s_at), score: 0.52 KIAA0586KIAA0586 (205631_at), score: 0.56 KIAA1024KIAA1024 (215081_at), score: -0.36 KIRRELkin of IRRE like (Drosophila) (220825_s_at), score: 0.59 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.48 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: -0.34 KNG1kininogen 1 (217512_at), score: -0.45 KRT14keratin 14 (209351_at), score: -0.36 KRT19keratin 19 (201650_at), score: -0.35 KRT2keratin 2 (207908_at), score: -0.43 KRT33Akeratin 33A (208483_x_at), score: -0.52 KTELC1KTEL (Lys-Tyr-Glu-Leu) containing 1 (218587_s_at), score: 0.43 LEMD3LEM domain containing 3 (218604_at), score: -0.42 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: -0.37 LIMK2LIM domain kinase 2 (202193_at), score: 0.42 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.53 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: 0.61 LOC100133558similar to hCG1642170 (216315_x_at), score: -0.34 LOC257152hypothetical protein LOC257152 (215302_at), score: -0.35 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.75 LOC390940similar to R28379_1 (213556_at), score: 0.49 LOC90379hypothetical protein BC002926 (221849_s_at), score: -0.39 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: -0.36 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (219346_at), score: 0.44 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: -0.45 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.34 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: 0.42 LRRC20leucine rich repeat containing 20 (218550_s_at), score: 0.5 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: -0.41 LUZP1leucine zipper protein 1 (221832_s_at), score: 0.45 LY75lymphocyte antigen 75 (205668_at), score: -0.4 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: -0.4 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.49 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.42 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.34 MBmyoglobin (204179_at), score: -0.37 MBD5methyl-CpG binding domain protein 5 (220195_at), score: 0.45 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: -0.37 MED13mediator complex subunit 13 (201986_at), score: 0.47 MEGF6multiple EGF-like-domains 6 (213942_at), score: -0.34 MGAMAX gene associated (212945_s_at), score: 0.51 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.41 MLPHmelanophilin (218211_s_at), score: 0.62 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.41 MMP24matrix metallopeptidase 24 (membrane-inserted) (49679_s_at), score: -0.37 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.54 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.54 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: 0.51 MTMR3myotubularin related protein 3 (202197_at), score: -0.38 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.55 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: -0.43 MYCL1v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) (214058_at), score: -0.41 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: 0.48 NAGAN-acetylgalactosaminidase, alpha- (202943_s_at), score: 0.48 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.51 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.35 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.35 NCAPH2non-SMC condensin II complex, subunit H2 (40640_at), score: 0.45 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.51 NEFLneurofilament, light polypeptide (221805_at), score: -0.5 NELFnasal embryonic LHRH factor (221214_s_at), score: -0.4 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.64 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.41 NFICnuclear factor I/C (CCAAT-binding transcription factor) (206929_s_at), score: 0.44 NMBneuromedin B (205204_at), score: 0.57 NOL8nucleolar protein 8 (218244_at), score: 0.62 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.44 NR0B1nuclear receptor subfamily 0, group B, member 1 (206644_at), score: -0.59 NR2C2nuclear receptor subfamily 2, group C, member 2 (206038_s_at), score: 0.52 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.34 NSD1nuclear receptor binding SET domain protein 1 (219084_at), score: 0.51 NSUN5BNOL1/NOP2/Sun domain family, member 5B (214100_x_at), score: 0.47 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.54 NUCKS1nuclear casein kinase and cyclin-dependent kinase substrate 1 (222027_at), score: 0.44 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.58 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.75 OLFM1olfactomedin 1 (213131_at), score: -0.37 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.43 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.38 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: -0.37 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.6 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.48 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.5 PDCphosducin (211496_s_at), score: -0.5 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.47 PEG3paternally expressed 3 (209242_at), score: -0.48 PENKproenkephalin (213791_at), score: -0.48 PF4V1platelet factor 4 variant 1 (207815_at), score: -0.38 PFDN6prefoldin subunit 6 (222029_x_at), score: 0.56 PFTK1PFTAIRE protein kinase 1 (204604_at), score: -0.37 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.47 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.56 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.43 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (212829_at), score: 0.43 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: -0.4 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.34 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.43 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: -0.34 PLGLB2plasminogen-like B2 (205871_at), score: -0.38 PLLPplasma membrane proteolipid (plasmolipin) (204519_s_at), score: -0.47 PLP1proteolipid protein 1 (210198_s_at), score: 0.46 PLXNB1plexin B1 (215807_s_at), score: -0.42 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: -0.51 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.42 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.43 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.39 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.51 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.43 PPFIBP1PTPRF interacting protein, binding protein 1 (liprin beta 1) (214374_s_at), score: 0.57 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: -0.36 PPM1Aprotein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform (210407_at), score: 0.58 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.35 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.69 PRR3proline rich 3 (204795_at), score: 0.43 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.48 PRSS3protease, serine, 3 (213421_x_at), score: -0.56 PSD3pleckstrin and Sec7 domain containing 3 (203355_s_at), score: 0.45 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.49 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.7 PTENP1phosphatase and tensin homolog pseudogene 1 (217492_s_at), score: 0.47 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.56 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.39 PTMAprothymosin, alpha (200772_x_at), score: 0.43 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.43 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.47 PXMP4peroxisomal membrane protein 4, 24kDa (219428_s_at), score: 0.45 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.47 QSOX1quiescin Q6 sulfhydryl oxidase 1 (201482_at), score: -0.38 RAB13RAB13, member RAS oncogene family (202252_at), score: 0.44 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.62 RABL3RAB, member of RAS oncogene family-like 3 (213970_at), score: 0.53 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.46 RALGDSral guanine nucleotide dissociation stimulator (209050_s_at), score: 0.53 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.36 RASA2RAS p21 protein activator 2 (206636_at), score: -0.48 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.43 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.46 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.46 RCC1regulator of chromosome condensation 1 (206499_s_at), score: 0.49 RFX7regulatory factor X, 7 (218430_s_at), score: 0.72 RHBDD3rhomboid domain containing 3 (204402_at), score: -0.42 RNF138ring finger protein 138 (218738_s_at), score: -0.36 RNF44ring finger protein 44 (203286_at), score: 0.58 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.39 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.38 RPL14ribosomal protein L14 (219138_at), score: 0.56 RPL27Aribosomal protein L27a (212044_s_at), score: 0.68 RPL38ribosomal protein L38 (202028_s_at), score: 0.52 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.45 RPS11ribosomal protein S11 (213350_at), score: 0.61 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.38 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.5 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.59 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: -0.46 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203789_s_at), score: -0.33 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.38 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.44 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.58 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: -0.35 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: -0.33 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.8 SEZ6L2seizure related 6 homolog (mouse)-like 2 (218720_x_at), score: -0.53 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.46 SIK2salt-inducible kinase 2 (213221_s_at), score: -0.33 SIPA1signal-induced proliferation-associated 1 (204164_at), score: -0.35 SIRPB1signal-regulatory protein beta 1 (206934_at), score: -0.53 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: 0.46 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.63 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.53 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: -0.4 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.35 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: -0.37 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: -0.47 SLC26A4solute carrier family 26, member 4 (206529_x_at), score: -0.49 SLC28A3solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 (220475_at), score: -0.47 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.65 SLC38A7solute carrier family 38, member 7 (218727_at), score: 0.45 SLKSTE20-like kinase (yeast) (206875_s_at), score: -0.41 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: 0.62 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.47 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.46 SNRPEsmall nuclear ribonucleoprotein polypeptide E (215450_at), score: 0.51 SNX13sorting nexin 13 (213292_s_at), score: 0.52 SORT1sortilin 1 (212807_s_at), score: 0.51 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: -0.34 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.38 SP140LSP140 nuclear body protein-like (214791_at), score: 0.45 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.54 SPAG6sperm associated antigen 6 (210033_s_at), score: -0.51 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.58 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: -0.44 SPHARS-phase response (cyclin-related) (206272_at), score: 0.42 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.5 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: -0.35 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.45 SRPRBsignal recognition particle receptor, B subunit (218140_x_at), score: 0.44 STC1stanniocalcin 1 (204597_x_at), score: -0.37 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.5 SYNE2spectrin repeat containing, nuclear envelope 2 (202761_s_at), score: 0.46 SYNMsynemin, intermediate filament protein (212730_at), score: 0.6 SYT1synaptotagmin I (203999_at), score: -0.54 SYT11synaptotagmin XI (209198_s_at), score: 0.63 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.48 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.35 TBL1Xtransducin (beta)-like 1X-linked (201867_s_at), score: 0.43 TBX5T-box 5 (207155_at), score: 0.44 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.48 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: 0.57 THEM2thioesterase superfamily member 2 (204565_at), score: 0.45 THOC2THO complex 2 (212994_at), score: 0.58 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.51 TMEM158transmembrane protein 158 (213338_at), score: -0.43 TMEM160transmembrane protein 160 (219219_at), score: 0.56 TMEM9BTMEM9 domain family, member B (218065_s_at), score: -0.34 TMSB15Athymosin beta 15a (205347_s_at), score: 0.63 TMUB2transmembrane and ubiquitin-like domain containing 2 (218419_s_at), score: 0.46 TNFRSF6Btumor necrosis factor receptor superfamily, member 6b, decoy (206467_x_at), score: 0.43 TNIKTRAF2 and NCK interacting kinase (213107_at), score: 0.44 TOMM22translocase of outer mitochondrial membrane 22 homolog (yeast) (217960_s_at), score: 0.56 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.45 TP53TG1TP53 target 1 (non-protein coding) (209917_s_at), score: 0.46 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.62 TPMTthiopurine S-methyltransferase (203672_x_at), score: 0.45 TRAPPC10trafficking protein particle complex 10 (209412_at), score: -0.36 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: 0.47 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: -0.35 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.48 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: 0.43 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: -0.45 TSPAN13tetraspanin 13 (217979_at), score: -0.34 TSPAN15tetraspanin 15 (218693_at), score: 0.45 TUBD1tubulin, delta 1 (210389_x_at), score: -0.33 TXNthioredoxin (216609_at), score: 0.47 TXNL4Bthioredoxin-like 4B (218794_s_at), score: 0.54 TYMPthymidine phosphorylase (217497_at), score: 0.44 UCHL5IPUCHL5 interacting protein (213334_x_at), score: 0.51 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 UIMC1ubiquitin interaction motif containing 1 (220746_s_at), score: 0.45 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.42 USE1unconventional SNARE in the ER 1 homolog (S. cerevisiae) (221706_s_at), score: 0.5 USP47ubiquitin specific peptidase 47 (221518_s_at), score: 0.53 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.59 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.34 VPS11vacuolar protein sorting 11 homolog (S. cerevisiae) (203292_s_at), score: 0.44 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.53 WDR43WD repeat domain 43 (214662_at), score: 0.51 WDR5BWD repeat domain 5B (219538_at), score: 0.47 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.44 XPO4exportin 4 (218479_s_at), score: 0.51 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.47 YY2YY2 transcription factor (216531_at), score: 0.59 ZC3H4zinc finger CCCH-type containing 4 (213390_at), score: 0.5 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.37 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: 0.55 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: 0.43 ZMAT5zinc finger, matrin type 5 (218752_at), score: 0.59 ZNF133zinc finger protein 133 (216960_s_at), score: 0.43 ZNF174zinc finger protein 174 (210291_s_at), score: 0.52 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.36 ZNF195zinc finger protein 195 (204234_s_at), score: 0.71 ZNF219zinc finger protein 219 (219314_s_at), score: -0.67 ZNF22zinc finger protein 22 (KOX 15) (218005_at), score: 0.57 ZNF223zinc finger protein 223 (207128_s_at), score: 0.51 ZNF304zinc finger protein 304 (207753_at), score: 0.57 ZNF34zinc finger protein 34 (219801_at), score: 0.45 ZNF350zinc finger protein 350 (219266_at), score: 0.54 ZNF415zinc finger protein 415 (205514_at), score: 0.42 ZNF434zinc finger protein 434 (218937_at), score: 0.48 ZNF536zinc finger protein 536 (206403_at), score: -0.34 ZNF667zinc finger protein 667 (207120_at), score: 0.67 ZNF688zinc finger protein 688 (213527_s_at), score: 0.52 ZNF749zinc finger protein 749 (215289_at), score: -0.44 ZNF93zinc finger protein 93 (208119_s_at), score: -0.36 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: 0.49 ZSCAN16zinc finger and SCAN domain containing 16 (219676_at), score: -0.36
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |