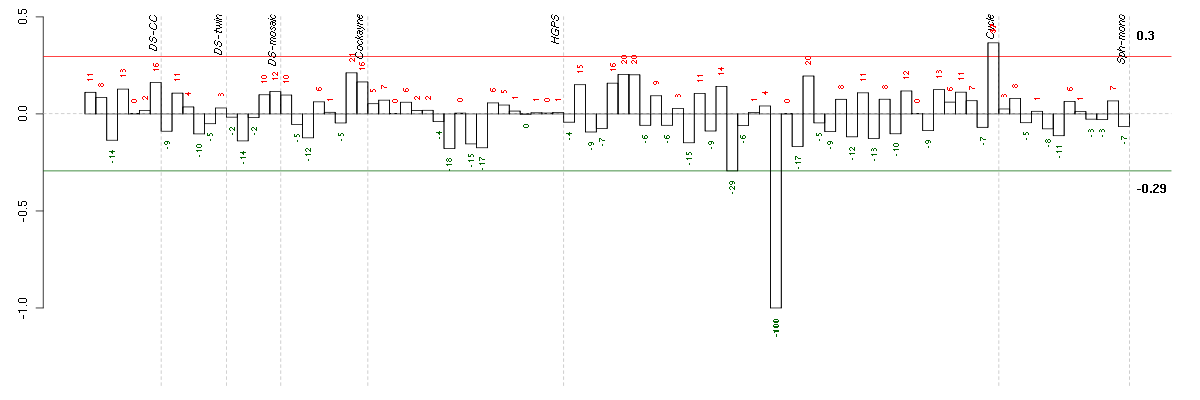

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signal transduction
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
regulation of G-protein coupled receptor protein signaling pathway
Any process that modulates the frequency, rate or extent of G-protein coupled receptor protein signaling pathway activity.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
negative regulation of signal transduction
Any process that stops, prevents or reduces the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
negative regulation of cell communication
Any process that decreases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
negative regulation of G-protein coupled receptor protein signaling pathway
Any process that stops, prevents or reduces the frequency, rate or extent of G-protein coupled receptor protein signaling pathway activity.
second-messenger-mediated signaling
A series of molecular signals in which an ion or small molecule is formed or released into the cytosol, thereby helping relay the signal within the cell.
cyclic-nucleotide-mediated signaling
A series of molecular signals in which a cell uses a cyclic nucleotide to convert an extracellular signal into a response.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of cell communication
Any process that decreases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
negative regulation of cell communication
Any process that decreases the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
negative regulation of signal transduction
Any process that stops, prevents or reduces the frequency, rate or extent of signal transduction.
negative regulation of signal transduction
Any process that stops, prevents or reduces the frequency, rate or extent of signal transduction.
regulation of G-protein coupled receptor protein signaling pathway
Any process that modulates the frequency, rate or extent of G-protein coupled receptor protein signaling pathway activity.
negative regulation of G-protein coupled receptor protein signaling pathway
Any process that stops, prevents or reduces the frequency, rate or extent of G-protein coupled receptor protein signaling pathway activity.
negative regulation of G-protein coupled receptor protein signaling pathway
Any process that stops, prevents or reduces the frequency, rate or extent of G-protein coupled receptor protein signaling pathway activity.
G-protein signaling, coupled to cyclic nucleotide second messenger
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand, followed by modulation of a nucleotide cyclase activity and a subsequent change in the concentration of a cyclic nucleotide.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
secretory granule
A small subcellular vesicle, surrounded by a membrane, that is formed from the Golgi apparatus and contains a highly concentrated protein destined for secretion. Secretory granules move towards the periphery of the cell, their membranes fuse with the cell membrane, and their protein load is exteriorized. Processing of the contained protein may take place in secretory granules.
secretory granule membrane
The lipid bilayer surrounding a secretory granule.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle membrane
The lipid bilayer surrounding any membrane-bounded vesicle in the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle part
Any constituent part of cytoplasmic vesicle, a vesicle formed of membrane or protein, found in the cytoplasm of a cell.
cytoplasmic vesicle membrane
The lipid bilayer surrounding a cytoplasmic vesicle.
secretory granule membrane
The lipid bilayer surrounding a secretory granule.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
amine binding
Interacting selectively with any organic compound that is weakly basic in character and contains an amino or a substituted amino group.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 4.244e-02 | 4.817 | 13 | 78 | Neuroactive ligand-receptor interaction |
ABHD6abhydrolase domain containing 6 (221679_s_at), score: 0.61 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.43 ACRV1acrosomal vesicle protein 1 (207969_x_at), score: 0.41 ACTN3actinin, alpha 3 (206891_at), score: 0.47 ADAadenosine deaminase (204639_at), score: 0.46 ADAM10ADAM metallopeptidase domain 10 (214895_s_at), score: 0.41 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.61 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.53 ADRB2adrenergic, beta-2-, receptor, surface (206170_at), score: -0.54 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.52 AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: -0.44 AHDC1AT hook, DNA binding motif, containing 1 (205002_at), score: -0.51 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: 0.43 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.45 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.46 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.49 ANGEL2angel homolog 2 (Drosophila) (221825_at), score: -0.45 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: 0.68 APOBEC3Aapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A (210873_x_at), score: -0.68 APOC1apolipoprotein C-I (213553_x_at), score: 0.42 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: -0.47 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: -0.57 ARHGAP10Rho GTPase activating protein 10 (219431_at), score: -0.63 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: -0.5 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: 0.44 ARHGEF15Rho guanine nucleotide exchange factor (GEF) 15 (217348_x_at), score: 0.58 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: -0.68 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.56 ASH1Lash1 (absent, small, or homeotic)-like (Drosophila) (218554_s_at), score: 0.49 ASPHaspartate beta-hydroxylase (205808_at), score: 0.59 ASTE1asteroid homolog 1 (Drosophila) (221135_s_at), score: -0.51 ATMataxia telangiectasia mutated (210858_x_at), score: 0.46 ATN1atrophin 1 (40489_at), score: -0.46 ATP13A3ATPase type 13A3 (219558_at), score: 0.69 ATP1B4ATPase, (Na+)/K+ transporting, beta 4 polypeptide (220556_at), score: 0.46 ATP6V1G2ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 (214762_at), score: 0.55 ATP8A1ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (213106_at), score: -0.56 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: 0.45 ATRNL1attractin-like 1 (213744_at), score: 0.55 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.44 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.52 BCAT1branched chain aminotransferase 1, cytosolic (214452_at), score: 0.54 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.51 BCL2L1BCL2-like 1 (215037_s_at), score: 0.46 BCL2L11BCL2-like 11 (apoptosis facilitator) (222343_at), score: 0.43 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: -0.65 BCORL1BCL6 co-repressor-like 1 (219444_at), score: -0.43 BCRbreakpoint cluster region (202315_s_at), score: -0.53 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.53 C10orf2chromosome 10 open reading frame 2 (218590_at), score: -0.46 C10orf28chromosome 10 open reading frame 28 (210455_at), score: 0.58 C11orf21chromosome 11 open reading frame 21 (220560_at), score: 0.54 C12orf51chromosome 12 open reading frame 51 (211034_s_at), score: -0.46 C13orf1chromosome 13 open reading frame 1 (214668_at), score: 0.43 C13orf34chromosome 13 open reading frame 34 (219544_at), score: -0.47 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: 0.48 C14orf93chromosome 14 open reading frame 93 (219009_at), score: 0.41 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.54 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.49 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.79 C1orf135chromosome 1 open reading frame 135 (220011_at), score: -0.46 C1orf156chromosome 1 open reading frame 156 (213528_at), score: -0.56 C1orf69chromosome 1 open reading frame 69 (215490_at), score: 0.46 C1QL1complement component 1, q subcomponent-like 1 (205575_at), score: 0.41 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.45 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.47 C2CD2LC2CD2-like (204757_s_at), score: -0.5 C2orf44chromosome 2 open reading frame 44 (219120_at), score: -0.51 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.46 C4orf30chromosome 4 open reading frame 30 (219717_at), score: -0.62 C4orf8chromosome 4 open reading frame 8 (203600_s_at), score: -0.43 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.46 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.71 C8orf17chromosome 8 open reading frame 17 (208266_at), score: 0.56 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.49 CA4carbonic anhydrase IV (206209_s_at), score: -0.42 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: 0.72 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.43 CAPN3calpain 3, (p94) (210944_s_at), score: 0.48 CBR4carbonyl reductase 4 (213626_at), score: -0.49 CCDC15coiled-coil domain containing 15 (220466_at), score: -0.48 CCDC46coiled-coil domain containing 46 (213644_at), score: -0.57 CCNFcyclin F (204826_at), score: -0.59 CCRN4LCCR4 carbon catabolite repression 4-like (S. cerevisiae) (220671_at), score: 0.67 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.49 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: 0.67 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.46 CDH8cadherin 8, type 2 (217574_at), score: -0.44 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.66 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.46 CENPOcentromere protein O (219472_at), score: 0.59 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: 0.48 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: 0.57 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: -0.56 CIDEBcell death-inducing DFFA-like effector b (221188_s_at), score: 0.51 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.54 CLCN2chloride channel 2 (213499_at), score: 0.66 CLDN10claudin 10 (205328_at), score: 0.43 CLIC4chloride intracellular channel 4 (201559_s_at), score: 0.62 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: 0.44 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: -0.44 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.52 CNR1cannabinoid receptor 1 (brain) (213436_at), score: -0.42 CNR2cannabinoid receptor 2 (macrophage) (206586_at), score: 0.55 COL10A1collagen, type X, alpha 1 (217428_s_at), score: -0.5 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.6 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.83 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.58 CPDcarboxypeptidase D (201942_s_at), score: 0.77 CPMcarboxypeptidase M (206100_at), score: -0.5 CRTC1CREB regulated transcription coactivator 1 (207159_x_at), score: -0.53 CRYBB2crystallin, beta B2 (206777_s_at), score: 0.51 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.45 CSKc-src tyrosine kinase (202329_at), score: -0.43 CTAGE5CTAGE family, member 5 (215930_s_at), score: 0.6 CTGLF1centaurin, gamma-like family, member 1 (221850_x_at), score: 0.61 CYBBcytochrome b-245, beta polypeptide (217431_x_at), score: -0.43 CYLC2cylicin, basic protein of sperm head cytoskeleton 2 (207780_at), score: 0.94 CYorf14chromosome Y open reading frame 14 (207063_at), score: -0.48 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (211440_x_at), score: 0.52 CYTH3cytohesin 3 (206523_at), score: 0.55 CYTH4cytohesin 4 (219183_s_at), score: 0.68 DAOD-amino-acid oxidase (206878_at), score: 0.67 DBPD site of albumin promoter (albumin D-box) binding protein (40273_at), score: -0.47 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.46 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.85 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: -0.49 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.44 DGKEdiacylglycerol kinase, epsilon 64kDa (207518_at), score: 0.45 DGKZdiacylglycerol kinase, zeta 104kDa (207556_s_at), score: -0.44 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.61 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.41 DISC1disrupted in schizophrenia 1 (206090_s_at), score: -0.54 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.46 DNAH7dynein, axonemal, heavy chain 7 (214222_at), score: 0.45 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.51 DRD2dopamine receptor D2 (206590_x_at), score: 0.57 DRD5dopamine receptor D5 (208486_at), score: 0.49 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.42 DTNAdystrobrevin, alpha (205741_s_at), score: 0.61 DUSP8dual specificity phosphatase 8 (206374_at), score: 0.55 DUSP9dual specificity phosphatase 9 (205777_at), score: 0.57 EDARectodysplasin A receptor (220048_at), score: 0.76 EDC4enhancer of mRNA decapping 4 (202496_at), score: -0.57 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: 0.66 EIF2B5eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa (212351_at), score: -0.43 ELAC1elaC homolog 1 (E. coli) (219325_s_at), score: 0.41 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.94 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: 0.42 ENO3enolase 3 (beta, muscle) (204483_at), score: 0.68 ENSAendosulfine alpha (221487_s_at), score: 0.53 ENTPD5ectonucleoside triphosphate diphosphohydrolase 5 (205757_at), score: -0.54 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.49 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: 0.45 ETV7ets variant 7 (221680_s_at), score: 0.45 F11coagulation factor XI (206610_s_at), score: 0.75 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: 0.53 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: -0.43 FAM3Afamily with sequence similarity 3, member A (38043_at), score: -0.46 FAM50Bfamily with sequence similarity 50, member B (205775_at), score: -0.53 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: 0.53 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.46 FGF12fibroblast growth factor 12 (207501_s_at), score: 0.44 FHITfragile histidine triad gene (206492_at), score: -0.5 FLCNfolliculin (215645_at), score: 0.62 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.42 FMO3flavin containing monooxygenase 3 (40665_at), score: -0.54 FMO4flavin containing monooxygenase 4 (206263_at), score: 0.42 FN1fibronectin 1 (214701_s_at), score: 0.73 FNDC4fibronectin type III domain containing 4 (218843_at), score: -0.46 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: 0.44 FOXN2forkhead box N2 (206708_at), score: 0.47 FRMD4BFERM domain containing 4B (213056_at), score: -0.53 FRS2fibroblast growth factor receptor substrate 2 (221308_at), score: 0.6 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.47 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.44 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.45 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: 0.54 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209893_s_at), score: -0.59 FZR1fizzy/cell division cycle 20 related 1 (Drosophila) (209416_s_at), score: -0.48 GABRG3gamma-aminobutyric acid (GABA) A receptor, gamma 3 (216895_at), score: -0.58 GANgigaxonin (220124_at), score: -0.65 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: -0.42 GATCglutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial) (214711_at), score: 0.8 GCS1glucosidase I (210627_s_at), score: -0.43 GFERgrowth factor, augmenter of liver regeneration (204659_s_at), score: -0.46 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: -0.51 GGNBP2gametogenetin binding protein 2 (218079_s_at), score: -0.45 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.55 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.48 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: 0.54 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: -0.72 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.7 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: 0.43 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.51 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: -0.47 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.46 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.8 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: -0.54 GPATCH4G patch domain containing 4 (220596_at), score: 0.52 GPATCH8G patch domain containing 8 (212485_at), score: -0.47 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.57 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: 0.49 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.72 HAB1B1 for mucin (215778_x_at), score: 0.4 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.49 HDAC7histone deacetylase 7 (217937_s_at), score: -0.49 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: 0.43 HFEhemochromatosis (206087_x_at), score: 0.45 HIST1H4Fhistone cluster 1, H4f (208026_at), score: -0.46 HLA-DMBmajor histocompatibility complex, class II, DM beta (203932_at), score: 0.45 HLA-DRB4major histocompatibility complex, class II, DR beta 4 (208306_x_at), score: 0.45 HLCSholocarboxylase synthetase (biotin-(proprionyl-Coenzyme A-carboxylase (ATP-hydrolysing)) ligase) (209399_at), score: 0.41 HN1Lhematological and neurological expressed 1-like (212109_at), score: -0.46 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.61 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: 0.5 HTATSF1HIV-1 Tat specific factor 1 (202601_s_at), score: 0.42 HTN1histatin 1 (206639_x_at), score: 0.45 HTR2B5-hydroxytryptamine (serotonin) receptor 2B (206638_at), score: -0.45 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.41 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.5 IFNA16interferon, alpha 16 (208448_x_at), score: 0.57 IFNAR1interferon (alpha, beta and omega) receptor 1 (204191_at), score: 0.45 IFNGR1interferon gamma receptor 1 (211676_s_at), score: 0.48 IFT88intraflagellar transport 88 homolog (Chlamydomonas) (204703_at), score: -0.47 IGFALSinsulin-like growth factor binding protein, acid labile subunit (215712_s_at), score: 0.46 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: 0.4 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.53 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: 0.41 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.56 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.45 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.58 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.41 IQCGIQ motif containing G (221185_s_at), score: -0.54 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.5 ITIH2inter-alpha (globulin) inhibitor H2 (204987_at), score: 0.48 JMJD4jumonji domain containing 4 (218560_s_at), score: -0.55 KAT2BK(lysine) acetyltransferase 2B (203845_at), score: -0.46 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.43 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.79 KIAA0317KIAA0317 (202128_at), score: -0.45 KIAA0649KIAA0649 (203955_at), score: -0.52 KIAA1009KIAA1009 (206005_s_at), score: 0.64 KITLGKIT ligand (211124_s_at), score: 0.81 KLF3Kruppel-like factor 3 (basic) (219657_s_at), score: 0.51 KRT24keratin 24 (220267_at), score: 0.45 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.46 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: 0.51 LARSleucyl-tRNA synthetase (217810_x_at), score: 0.47 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: 0.58 LBRlamin B receptor (201795_at), score: -0.45 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: 0.5 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.43 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.74 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: 0.73 LOC100129502hypothetical protein LOC100129502 (217548_at), score: 0.56 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.59 LOC100133748similar to GTF2IRD2 protein (215569_at), score: 0.55 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: 0.5 LOC285412similar to KIAA0737 protein (217448_s_at), score: 0.56 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: 0.48 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (214712_at), score: -0.91 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.72 LOC81691exonuclease NEF-sp (208107_s_at), score: -0.65 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: -0.45 LOXlysyl oxidase (213640_s_at), score: 0.44 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.41 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.51 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: -0.43 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.91 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: 0.44 MADDMAP-kinase activating death domain (38398_at), score: -0.6 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: 0.7 MAN2B1mannosidase, alpha, class 2B, member 1 (209166_s_at), score: -0.55 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.48 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.46 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: 0.45 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: -0.44 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.46 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: -0.77 MAXMYC associated factor X (210734_x_at), score: 0.7 MBmyoglobin (204179_at), score: -0.44 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: 0.7 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: 0.61 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: -0.73 MDM2Mdm2 p53 binding protein homolog (mouse) (217373_x_at), score: 0.57 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: 0.54 MEGF8multiple EGF-like-domains 8 (214778_at), score: -0.45 MEP1Bmeprin A, beta (207251_at), score: 0.4 METmet proto-oncogene (hepatocyte growth factor receptor) (211599_x_at), score: 0.63 MGST2microsomal glutathione S-transferase 2 (204168_at), score: -0.49 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.43 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: -0.48 MOAP1modulator of apoptosis 1 (212508_at), score: 0.44 MR1major histocompatibility complex, class I-related (207565_s_at), score: 0.4 MRPL2mitochondrial ribosomal protein L2 (218887_at), score: -0.43 MTRF1mitochondrial translational release factor 1 (219822_at), score: -0.47 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.49 MYL10myosin, light chain 10, regulatory (221659_s_at), score: 0.45 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.41 MYO15Amyosin XVA (220288_at), score: -0.44 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: -0.58 NAALAD2N-acetylated alpha-linked acidic dipeptidase 2 (222161_at), score: -0.59 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.41 NDE1nudE nuclear distribution gene E homolog 1 (A. nidulans) (218414_s_at), score: -0.52 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: -0.44 NF1neurofibromin 1 (211094_s_at), score: 0.5 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: 0.43 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.42 NMNAT2nicotinamide nucleotide adenylyltransferase 2 (209755_at), score: -0.6 NOC4Lnucleolar complex associated 4 homolog (S. cerevisiae) (218860_at), score: -0.56 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.44 NPAL2NIPA-like domain containing 2 (220128_s_at), score: 0.65 NPHP4nephronophthisis 4 (213471_at), score: -0.5 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.41 NR1D2nuclear receptor subfamily 1, group D, member 2 (209750_at), score: 0.43 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.48 NUP93nucleoporin 93kDa (202188_at), score: -0.45 OLAHoleoyl-ACP hydrolase (219975_x_at), score: -0.45 OPRL1opiate receptor-like 1 (206564_at), score: 0.63 OR1E1olfactory receptor, family 1, subfamily E, member 1 (214515_at), score: 0.62 OR2C1olfactory receptor, family 2, subfamily C, member 1 (221460_at), score: 0.68 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: 0.47 OSMRoncostatin M receptor (205729_at), score: 0.77 OTUB2OTU domain, ubiquitin aldehyde binding 2 (219369_s_at), score: 0.45 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.55 PAPOLApoly(A) polymerase alpha (212720_at), score: 0.44 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.48 PCDH24protocadherin 24 (220186_s_at), score: 0.42 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.67 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.66 PCDHGB5protocadherin gamma subfamily B, 5 (211807_x_at), score: 0.42 PCF11PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae) (203378_at), score: -0.65 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: -0.99 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.5 PEX11Aperoxisomal biogenesis factor 11 alpha (205160_at), score: -0.63 PFDN6prefoldin subunit 6 (222029_x_at), score: 0.44 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: 0.49 PHF16PHD finger protein 16 (204866_at), score: -0.56 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.78 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: 0.45 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (204144_s_at), score: -0.43 PIK3R5phosphoinositide-3-kinase, regulatory subunit 5 (220566_at), score: 0.56 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.52 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.44 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.44 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: 0.48 PLA2R1phospholipase A2 receptor 1, 180kDa (207415_at), score: -0.44 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: -0.53 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.41 PLXNA2plexin A2 (213030_s_at), score: -0.48 PMP2peripheral myelin protein 2 (206826_at), score: -0.69 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: 0.41 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.44 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.53 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (217610_at), score: 0.62 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: -0.55 POLSpolymerase (DNA directed) sigma (202466_at), score: -0.55 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.47 POMT1protein-O-mannosyltransferase 1 (218476_at), score: -0.55 PPCDCphosphopantothenoylcysteine decarboxylase (219066_at), score: -0.69 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha isoform (202187_s_at), score: -0.51 PPP2R5Bprotein phosphatase 2, regulatory subunit B', beta isoform (635_s_at), score: -0.43 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.46 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: -0.48 PRF1perforin 1 (pore forming protein) (214617_at), score: 0.49 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.75 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (214474_at), score: 0.41 PRKCDprotein kinase C, delta (202545_at), score: -0.75 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.47 PRR14proline rich 14 (218714_at), score: -0.51 PRR4proline rich 4 (lacrimal) (204919_at), score: 0.72 PRRX1paired related homeobox 1 (205991_s_at), score: 0.52 PRSS12protease, serine, 12 (neurotrypsin, motopsin) (205515_at), score: 0.43 PRSS22protease, serine, 22 (205847_at), score: 0.42 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.43 PTCH1patched homolog 1 (Drosophila) (209815_at), score: 0.49 PTH1Rparathyroid hormone 1 receptor (205911_at), score: 0.42 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.58 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.45 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.5 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.56 PTPRUprotein tyrosine phosphatase, receptor type, U (211320_s_at), score: 0.6 PXMP4peroxisomal membrane protein 4, 24kDa (219428_s_at), score: -0.47 RAB35RAB35, member RAS oncogene family (205461_at), score: 0.78 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.69 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.54 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: 0.42 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.76 RANBP10RAN binding protein 10 (221809_at), score: -0.78 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: -0.44 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (219112_at), score: -0.72 RASSF9Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 (210335_at), score: 0.51 RBBP9retinoblastoma binding protein 9 (221440_s_at), score: 0.83 RBM38RNA binding motif protein 38 (212430_at), score: 0.53 RCAN1regulator of calcineurin 1 (215253_s_at), score: 0.44 RCOR3REST corepressor 3 (218344_s_at), score: 0.41 RDXradixin (204969_s_at), score: 0.44 RECQL5RecQ protein-like 5 (34063_at), score: 0.44 REEP4receptor accessory protein 4 (218777_at), score: -0.47 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: 0.41 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.64 RNASEH2Bribonuclease H2, subunit B (219056_at), score: -0.45 RNF128ring finger protein 128 (219263_at), score: -0.72 RNF19Aring finger protein 19A (220483_s_at), score: 0.59 RNF19Bring finger protein 19B (213038_at), score: -0.43 RNF6ring finger protein (C3H2C3 type) 6 (210932_s_at), score: 0.47 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: 0.46 RNGTTRNA guanylyltransferase and 5'-phosphatase (204207_s_at), score: 0.44 RP11-138L21.1similar to cell recognition molecule CASPR3 (220436_at), score: -0.58 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.5 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.4 RPS20ribosomal protein S20 (216247_at), score: 0.44 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.48 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: 0.62 RRP7Bribosomal RNA processing 7 homolog B (S. cerevisiae) (214828_s_at), score: 0.44 RRP8ribosomal RNA processing 8, methyltransferase, homolog (yeast) (203171_s_at), score: -0.43 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.53 RXRBretinoid X receptor, beta (215099_s_at), score: 0.41 S100A14S100 calcium binding protein A14 (218677_at), score: 0.44 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.59 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: 0.54 SCAND2SCAN domain containing 2 pseudogene (222177_s_at), score: -0.53 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: 0.51 SCYL2SCY1-like 2 (S. cerevisiae) (221220_s_at), score: 0.45 SERGEFsecretion regulating guanine nucleotide exchange factor (220482_s_at), score: -0.52 SFRS1splicing factor, arginine/serine-rich 1 (201742_x_at), score: 0.42 SFRS16splicing factor, arginine/serine-rich 16 (204978_at), score: 0.54 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.66 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: 0.48 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: -0.44 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.54 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.75 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: -0.47 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.45 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (203727_at), score: -0.56 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.53 SLC14A1solute carrier family 14 (urea transporter), member 1 (Kidd blood group) (205856_at), score: -0.63 SLC22A14solute carrier family 22, member 14 (207408_at), score: 0.41 SLC25A21solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 (220474_at), score: -0.69 SLC25A40solute carrier family 25, member 40 (205716_at), score: 0.49 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.59 SLC35F5solute carrier family 35, member F5 (220123_at), score: -0.47 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.48 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.73 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.49 SMURF1SMAD specific E3 ubiquitin protein ligase 1 (212666_at), score: -0.47 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.63 SNTG1syntrophin, gamma 1 (220405_at), score: -0.54 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.44 SP140SP140 nuclear body protein (207777_s_at), score: 0.47 SPATA6spermatogenesis associated 6 (220298_s_at), score: 0.56 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.43 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.56 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.43 SSTR1somatostatin receptor 1 (208482_at), score: 0.51 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.56 STRA6stimulated by retinoic acid gene 6 homolog (mouse) (221701_s_at), score: -0.48 STRNstriatin, calmodulin binding protein (205520_at), score: 0.43 SYPL1synaptophysin-like 1 (201259_s_at), score: 0.54 TAC3tachykinin 3 (219992_at), score: 0.64 TAF4TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa (213090_s_at), score: -0.69 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.58 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.43 TAS2R14taste receptor, type 2, member 14 (221391_at), score: 0.42 TAS2R9taste receptor, type 2, member 9 (221461_at), score: 0.51 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: -0.44 TBL3transducin (beta)-like 3 (209820_s_at), score: -0.82 TBPTATA box binding protein (203135_at), score: -0.46 TBX10T-box 10 (207689_at), score: 0.43 TBX21T-box 21 (220684_at), score: 0.8 TCEB3Btranscription elongation factor B polypeptide 3B (elongin A2) (220844_at), score: -0.56 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (34260_at), score: -0.56 TEP1telomerase-associated protein 1 (205727_at), score: 0.51 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.61 TMEM187transmembrane protein 187 (204340_at), score: 0.41 TMEM209transmembrane protein 209 (222267_at), score: 0.42 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.46 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.48 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: 0.59 TOB2transducer of ERBB2, 2 (221496_s_at), score: -0.44 TRAK1trafficking protein, kinesin binding 1 (214924_s_at), score: -0.43 TRAM1translocation associated membrane protein 1 (210733_at), score: 0.47 TRIM25tripartite motif-containing 25 (206911_at), score: 0.54 TRMT2BTRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae) (205238_at), score: 0.49 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: 0.49 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: 0.47 TSHBthyroid stimulating hormone, beta (214529_at), score: 0.43 TTLL7tubulin tyrosine ligase-like family, member 7 (219882_at), score: 0.49 UBR2ubiquitin protein ligase E3 component n-recognin 2 (212760_at), score: -0.44 UBXN4UBX domain protein 4 (212008_at), score: 0.5 UGCGL1UDP-glucose ceramide glucosyltransferase-like 1 (218257_s_at), score: 0.45 USP34ubiquitin specific peptidase 34 (212065_s_at), score: 0.43 UTRNutrophin (213022_s_at), score: 0.61 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: -0.55 VCLvinculin (200930_s_at), score: 0.4 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.44 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.45 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.82 WDR78WD repeat domain 78 (220769_s_at), score: 0.56 WFDC2WAP four-disulfide core domain 2 (203892_at), score: 0.64 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: 0.43 WNT10Bwingless-type MMTV integration site family, member 10B (206213_at), score: 0.44 XAB2XPA binding protein 2 (218110_at), score: -0.56 XPO4exportin 4 (218479_s_at), score: 0.42 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: 0.42 ZDHHC13zinc finger, DHHC-type containing 13 (219296_at), score: -0.61 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.67 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: 0.53 ZFP37zinc finger protein 37 homolog (mouse) (207068_at), score: 0.78 ZMYND11zinc finger, MYND domain containing 11 (202137_s_at), score: 0.43 ZNF117zinc finger protein 117 (207605_x_at), score: 0.47 ZNF132zinc finger protein 132 (207402_at), score: -0.52 ZNF137zinc finger protein 137 (207394_at), score: -0.75 ZNF224zinc finger protein 224 (216983_s_at), score: 0.5 ZNF236zinc finger protein 236 (47571_at), score: -1 ZNF26zinc finger protein 26 (219595_at), score: -0.53 ZNF280Azinc finger protein 280A (216034_at), score: -0.43 ZNF37Bzinc finger protein 37B (215358_x_at), score: 0.43 ZNF443zinc finger protein 443 (205928_at), score: -0.57 ZNF460zinc finger protein 460 (216279_at), score: -0.44 ZNF467zinc finger protein 467 (214746_s_at), score: 0.47 ZNF510zinc finger protein 510 (206053_at), score: 0.42 ZNF529zinc finger protein 529 (215307_at), score: 0.51 ZNF571zinc finger protein 571 (206648_at), score: -0.5 ZNF574zinc finger protein 574 (218762_at), score: -0.74 ZNF589zinc finger protein 589 (210062_s_at), score: 0.48 ZNF606zinc finger protein 606 (219635_at), score: -0.44 ZNF614zinc finger protein 614 (220721_at), score: 0.55 ZNF665zinc finger protein 665 (220760_x_at), score: 0.53 ZNF669zinc finger protein 669 (220215_at), score: 0.55 ZNF675zinc finger protein 675 (217547_x_at), score: 0.48 ZNF692zinc finger protein 692 (220661_s_at), score: -0.44 ZRSR2zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 (213876_x_at), score: -0.51 ZSCAN16zinc finger and SCAN domain containing 16 (219676_at), score: -0.79 ZXDAzinc finger, X-linked, duplicated A (215263_at), score: 0.46
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |