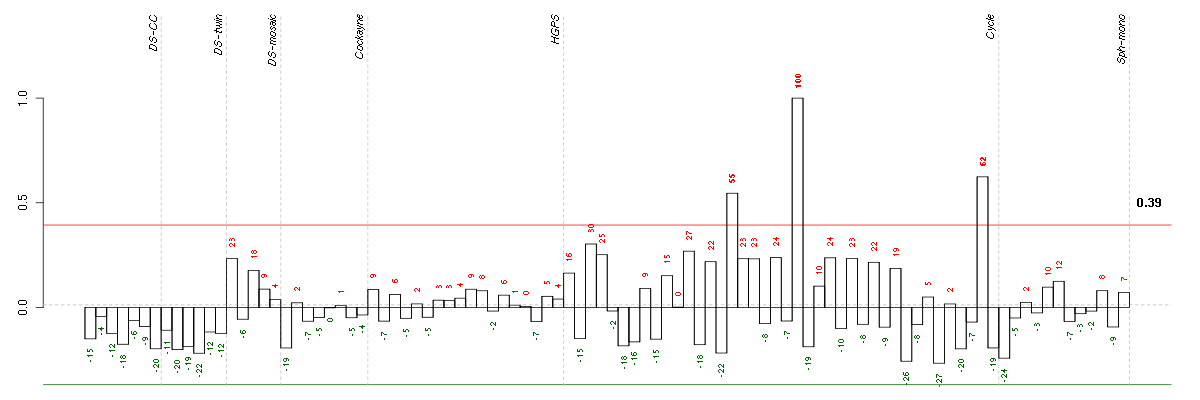

Under-expression is coded with green,
over-expression with red color.

cell activation
A change in the morphology or behavior of a cell resulting from exposure to an activating factor such as a cellular or soluble ligand.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
all
This term is the most general term possible


A1CFAPOBEC1 complementation factor (220951_s_at), score: -0.57 AASSaminoadipate-semialdehyde synthase (214829_at), score: -0.45 ABCG2ATP-binding cassette, sub-family G (WHITE), member 2 (209735_at), score: 0.53 ACAD10acyl-Coenzyme A dehydrogenase family, member 10 (219986_s_at), score: -0.73 ACRV1acrosomal vesicle protein 1 (207969_x_at), score: -0.57 ACVR1Bactivin A receptor, type IB (213198_at), score: -0.59 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: -0.45 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: -0.47 ADARB1adenosine deaminase, RNA-specific, B1 (RED1 homolog rat) (203865_s_at), score: -0.47 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.7 ADRB2adrenergic, beta-2-, receptor, surface (206170_at), score: 0.74 AFF1AF4/FMR2 family, member 1 (201924_at), score: -0.51 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: 0.49 AMACRalpha-methylacyl-CoA racemase (209424_s_at), score: 0.48 AMELXamelogenin (amelogenesis imperfecta 1, X-linked) (208410_x_at), score: -0.55 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: -0.57 ANK1ankyrin 1, erythrocytic (208353_x_at), score: -0.63 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: 0.68 APOC4apolipoprotein C-IV (206738_at), score: -0.53 APOL1apolipoprotein L, 1 (209546_s_at), score: -0.5 ARandrogen receptor (211621_at), score: -0.58 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: -0.63 ARHGEF15Rho guanine nucleotide exchange factor (GEF) 15 (217348_x_at), score: -0.72 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (218501_at), score: -0.58 ARID3AAT rich interactive domain 3A (BRIGHT-like) (205865_at), score: -0.45 ARID4BAT rich interactive domain 4B (RBP1-like) (221230_s_at), score: -0.55 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: 0.53 ATF4activating transcription factor 4 (tax-responsive enhancer element B67) (200779_at), score: -0.46 ATF5activating transcription factor 5 (204999_s_at), score: 0.46 ATF6Bactivating transcription factor 6 beta (203168_at), score: -0.47 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: -0.48 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: -0.53 ATP6V1G2ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 (214762_at), score: -0.47 ATXN1ataxin 1 (203232_s_at), score: -0.67 AVL9AVL9 homolog (S. cerevisiase) (212474_at), score: -0.48 AVPI1arginine vasopressin-induced 1 (218631_at), score: -0.58 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.69 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.49 BCANbrevican (91920_at), score: -0.62 BCL6B-cell CLL/lymphoma 6 (203140_at), score: -0.47 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.51 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.49 BMP10bone morphogenetic protein 10 (208292_at), score: -0.76 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: -0.48 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.73 C10orf110chromosome 10 open reading frame 110 (220703_at), score: -0.51 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.48 C10orf81chromosome 10 open reading frame 81 (219857_at), score: 0.77 C12orf51chromosome 12 open reading frame 51 (211034_s_at), score: -0.62 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.49 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.55 C1orf61chromosome 1 open reading frame 61 (205103_at), score: -0.73 C1orf66chromosome 1 open reading frame 66 (218914_at), score: -0.48 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.66 C20orf11chromosome 20 open reading frame 11 (218448_at), score: 0.5 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.62 C4orf16chromosome 4 open reading frame 16 (219023_at), score: -0.56 C6orf155chromosome 6 open reading frame 155 (220324_at), score: 0.57 C6orf162chromosome 6 open reading frame 162 (213314_at), score: -0.52 C6orf47chromosome 6 open reading frame 47 (204968_at), score: 0.46 C7orf44chromosome 7 open reading frame 44 (209445_x_at), score: 0.45 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.49 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.67 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.54 C9orf167chromosome 9 open reading frame 167 (219620_x_at), score: 0.45 C9orf86chromosome 9 open reading frame 86 (219828_at), score: 0.65 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.76 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.45 CBX4chromobox homolog 4 (Pc class homolog, Drosophila) (206724_at), score: 0.57 CCDC15coiled-coil domain containing 15 (220466_at), score: 0.52 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.59 CD22CD22 molecule (38521_at), score: -0.45 CD28CD28 molecule (206545_at), score: 0.46 CD53CD53 molecule (203416_at), score: -0.48 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: -0.55 CDC23cell division cycle 23 homolog (S. cerevisiae) (202892_at), score: -0.46 CDCA4cell division cycle associated 4 (218399_s_at), score: 0.54 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: -0.57 CFHR2complement factor H-related 2 (206910_x_at), score: -0.59 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.45 CHRDL1chordin-like 1 (209763_at), score: -0.48 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: 0.48 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (219634_at), score: 0.47 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.53 CLCN6chloride channel 6 (203950_s_at), score: -0.55 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: -0.46 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (219340_s_at), score: -0.5 CNKSR1connector enhancer of kinase suppressor of Ras 1 (204740_at), score: -0.47 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.56 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.49 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: -0.46 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.56 COX16COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) (217645_at), score: -0.53 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: -0.48 CPEB1cytoplasmic polyadenylation element binding protein 1 (219578_s_at), score: -0.54 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: 0.7 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: -0.55 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: 0.63 CSNK1G1casein kinase 1, gamma 1 (221673_s_at), score: 0.53 CTHcystathionase (cystathionine gamma-lyase) (217127_at), score: -0.61 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.56 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.53 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: -0.49 DCLRE1BDNA cross-link repair 1B (PSO2 homolog, S. cerevisiae) (219490_s_at), score: 0.56 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: -0.51 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: 0.62 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: -0.47 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.59 DGKIdiacylglycerol kinase, iota (206806_at), score: 0.64 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: 0.46 DNAJB2DnaJ (Hsp40) homolog, subfamily B, member 2 (202500_at), score: -0.5 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.58 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.5 DOK4docking protein 4 (209691_s_at), score: 0.49 DOPEY1dopey family member 1 (40612_at), score: -0.49 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: -0.52 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.75 DUSP8dual specificity phosphatase 8 (206374_at), score: -0.64 DXS542X-linked retinopathy protein-like (222247_at), score: -0.59 EDARectodysplasin A receptor (220048_at), score: -0.6 EFHC1EF-hand domain (C-terminal) containing 1 (219833_s_at), score: 0.53 EFNB1ephrin-B1 (202711_at), score: 0.49 EHHADHenoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase (205222_at), score: -0.56 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: 0.46 ELA3Aelastase 3A, pancreatic (211738_x_at), score: -0.53 ENGASEendo-beta-N-acetylglucosaminidase (220349_s_at), score: -0.49 ENO3enolase 3 (beta, muscle) (204483_at), score: -0.67 ENOX2ecto-NOX disulfide-thiol exchanger 2 (32042_at), score: -0.69 EPB41L5erythrocyte membrane protein band 4.1 like 5 (220977_x_at), score: -0.65 EPHA3EPH receptor A3 (211164_at), score: 0.47 ERMAPerythroblast membrane-associated protein (Scianna blood group) (219905_at), score: -0.69 EVLEnah/Vasp-like (217838_s_at), score: 0.54 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: -0.5 FAM118Afamily with sequence similarity 118, member A (219629_at), score: -0.48 FBRSfibrosin (218255_s_at), score: -0.63 FBXL6F-box and leucine-rich repeat protein 6 (219189_at), score: 0.52 FBXO11F-box protein 11 (203255_at), score: -0.44 FBXO2F-box protein 2 (219305_x_at), score: -0.63 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.8 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.46 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.45 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: -0.51 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: -0.61 FGL1fibrinogen-like 1 (205305_at), score: -0.6 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.62 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.47 FLJ20674hypothetical protein FLJ20674 (220137_at), score: -0.52 FLJ21369hypothetical protein FLJ21369 (220401_at), score: -0.45 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.63 FLRT1fibronectin leucine rich transmembrane protein 1 (210414_at), score: -0.45 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.5 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: -0.48 GABRA5gamma-aminobutyric acid (GABA) A receptor, alpha 5 (206456_at), score: -0.46 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.48 GFERgrowth factor, augmenter of liver regeneration (204659_s_at), score: 0.55 GFOD2glucose-fructose oxidoreductase domain containing 2 (221028_s_at), score: 0.46 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.48 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (206355_at), score: 0.61 GNAZguanine nucleotide binding protein (G protein), alpha z polypeptide (204993_at), score: -0.5 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: -0.47 GOLGA9Pgolgi autoantigen, golgin subfamily a, 9 pseudogene (213737_x_at), score: -0.59 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: -0.61 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.5 GP6glycoprotein VI (platelet) (220336_s_at), score: 0.81 GPR144G protein-coupled receptor 144 (216289_at), score: -0.74 GRB10growth factor receptor-bound protein 10 (210999_s_at), score: -0.45 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.52 GSTA1glutathione S-transferase alpha 1 (215766_at), score: -0.53 GTPBP1GTP binding protein 1 (219357_at), score: 0.6 GVIN1GTPase, very large interferon inducible 1 (220577_at), score: -0.58 HAB1B1 for mucin (215778_x_at), score: -0.46 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.47 HBBhemoglobin, beta (211696_x_at), score: -0.59 HCRP1hepatocellular carcinoma-related HCRP1 (216176_at), score: -0.51 HDAC4histone deacetylase 4 (204225_at), score: -0.63 HDAC9histone deacetylase 9 (205659_at), score: 0.63 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: -0.56 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.57 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.51 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.47 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.49 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.52 HOOK1hook homolog 1 (Drosophila) (219976_at), score: -0.71 HOXB5homeobox B5 (205601_s_at), score: -0.58 HPS4Hermansky-Pudlak syndrome 4 (54037_at), score: -0.5 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: -0.48 HTN1histatin 1 (206639_x_at), score: -0.48 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.64 IFI44Linterferon-induced protein 44-like (204439_at), score: -0.51 IFNGinterferon, gamma (210354_at), score: -0.5 IFT74intraflagellar transport 74 homolog (Chlamydomonas) (219174_at), score: -0.44 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (207901_at), score: -0.48 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.59 IL7interleukin 7 (206693_at), score: -0.5 ING4inhibitor of growth family, member 4 (48825_at), score: -0.52 INHAinhibin, alpha (210141_s_at), score: 0.48 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.51 IPO8importin 8 (205701_at), score: -0.46 ISOC2isochorismatase domain containing 2 (218893_at), score: -0.47 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.51 JMJD2Cjumonji domain containing 2C (209984_at), score: -0.55 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.5 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.45 KIAA0240KIAA0240 (38892_at), score: -0.64 KIAA0644KIAA0644 gene product (205150_s_at), score: 0.52 KIAA0894KIAA0894 protein (207436_x_at), score: -0.51 KIAA0947KIAA0947 protein (209654_at), score: -0.46 KIF13Bkinesin family member 13B (202962_at), score: -0.49 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: -0.48 KNG1kininogen 1 (217512_at), score: -0.45 KPNA5karyopherin alpha 5 (importin alpha 6) (206241_at), score: -0.48 KRT14keratin 14 (209351_at), score: 0.48 KRT24keratin 24 (220267_at), score: -0.45 KRT33Akeratin 33A (208483_x_at), score: 0.61 KRT86keratin 86 (215189_at), score: -0.62 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: 0.91 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.53 LMBR1Llimb region 1 homolog (mouse)-like (220036_s_at), score: -0.58 LMF2lipase maturation factor 2 (212682_s_at), score: 0.48 LOC100133503similar to LRRC37A3 protein (220219_s_at), score: -0.65 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.51 LOC149501similar to keratin 8 (216821_at), score: -0.48 LOC257152hypothetical protein LOC257152 (215302_at), score: 0.45 LOC729806similar to hCG1725380 (217544_at), score: -0.45 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (214712_at), score: -0.45 LOC80054hypothetical LOC80054 (220465_at), score: -0.48 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.53 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: 0.46 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.6 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: -0.62 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: 0.55 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.47 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.68 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.58 MAP3K7IP1mitogen-activated protein kinase kinase kinase 7 interacting protein 1 (203901_at), score: -0.51 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.6 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: -0.46 MAPRE3microtubule-associated protein, RP/EB family, member 3 (214270_s_at), score: 0.47 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: 0.45 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.72 MATN3matrilin 3 (206091_at), score: 0.76 MBmyoglobin (204179_at), score: 0.49 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: -0.51 MCM9minichromosome maintenance complex component 9 (219673_at), score: -0.51 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: -0.5 MEP1Bmeprin A, beta (207251_at), score: 0.77 MERTKc-mer proto-oncogene tyrosine kinase (211913_s_at), score: -0.51 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: 0.49 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.46 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.81 MNTMAX binding protein (204206_at), score: -0.69 MSX2msh homeobox 2 (210319_x_at), score: -0.48 MTM1myotubularin 1 (36920_at), score: -0.45 MTMR4myotubularin related protein 4 (212277_at), score: -0.48 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.5 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.54 MUM1melanoma associated antigen (mutated) 1 (221290_s_at), score: -0.48 MYO1Amyosin IA (211916_s_at), score: -0.47 NCAM1neural cell adhesion molecule 1 (212843_at), score: -0.48 NCAM2neural cell adhesion molecule 2 (205669_at), score: -0.69 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: -0.5 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.45 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.53 NCRNA00153non-protein coding RNA 153 (219961_s_at), score: -0.47 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.45 NFIXnuclear factor I/X (CCAAT-binding transcription factor) (209807_s_at), score: -0.64 NINninein (GSK3B interacting protein) (219285_s_at), score: 0.5 NOL10nucleolar protein 10 (218591_s_at), score: -0.46 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.58 NR1D2nuclear receptor subfamily 1, group D, member 2 (209750_at), score: -0.62 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: -0.49 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.61 OCLMoculomedin (208274_at), score: -0.46 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: -0.52 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.49 OGTO-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) (209240_at), score: -0.54 OPRL1opiate receptor-like 1 (206564_at), score: -0.53 OR2B2olfactory receptor, family 2, subfamily B, member 2 (216408_at), score: -0.52 OR3A2olfactory receptor, family 3, subfamily A, member 2 (221386_at), score: -0.51 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.5 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.57 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: -0.57 P2RY4pyrimidinergic receptor P2Y, G-protein coupled, 4 (221466_at), score: -0.61 PAPOLGpoly(A) polymerase gamma (222273_at), score: -0.51 PARP11poly (ADP-ribose) polymerase family, member 11 (220315_at), score: -0.47 PARVBparvin, beta (37965_at), score: 0.49 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: -0.57 PDE12phosphodiesterase 12 (214826_at), score: -0.57 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: -0.47 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.45 PER3period homolog 3 (Drosophila) (221045_s_at), score: -0.56 PEX16peroxisomal biogenesis factor 16 (221604_s_at), score: 0.46 PFDN6prefoldin subunit 6 (222029_x_at), score: -0.47 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.51 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: -0.52 PHF21APHD finger protein 21A (203278_s_at), score: -0.54 PHF7PHD finger protein 7 (215622_x_at), score: -0.48 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: -0.47 PIGCphosphatidylinositol glycan anchor biosynthesis, class C (216593_s_at), score: 0.54 PIK3CBphosphoinositide-3-kinase, catalytic, beta polypeptide (212688_at), score: -0.49 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: -0.61 PIP5K1Bphosphatidylinositol-4-phosphate 5-kinase, type I, beta (217477_at), score: 0.71 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: -0.94 PISDphosphatidylserine decarboxylase (202392_s_at), score: 0.64 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.56 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.45 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (204613_at), score: -0.56 PLP1proteolipid protein 1 (210198_s_at), score: -0.45 POLEpolymerase (DNA directed), epsilon (216026_s_at), score: 0.6 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (205218_at), score: 0.47 PPP1R10protein phosphatase 1, regulatory (inhibitor) subunit 10 (201702_s_at), score: 0.55 PPT2palmitoyl-protein thioesterase 2 (209490_s_at), score: 0.45 PRAMEF11PRAME family member 11 (217365_at), score: -0.67 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: -0.59 PRDM10PR domain containing 10 (219515_at), score: -0.52 PRKCBprotein kinase C, beta (209685_s_at), score: -0.57 PRKCDprotein kinase C, delta (202545_at), score: 0.81 PRKYprotein kinase, Y-linked (206279_at), score: -0.6 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.57 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.75 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: -0.54 PXNpaxillin (211823_s_at), score: 0.57 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.76 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.49 RAB15RAB15, member RAS onocogene family (221810_at), score: 0.55 RAB33ARAB33A, member RAS oncogene family (206039_at), score: -0.51 RAB40BRAB40B, member RAS oncogene family (204547_at), score: 0.63 RABIFRAB interacting factor (204478_s_at), score: 0.47 RANBP3RAN binding protein 3 (210120_s_at), score: 0.57 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.48 RASSF9Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 (210335_at), score: -0.48 RBBP6retinoblastoma binding protein 6 (212781_at), score: -0.75 RBMS3RNA binding motif, single stranded interacting protein (206767_at), score: -0.46 RCL1RNA terminal phosphate cyclase-like 1 (218544_s_at), score: -0.57 RFX3regulatory factor X, 3 (influences HLA class II expression) (207234_at), score: 1 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: -0.63 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: -0.47 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.64 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.62 RLFrearranged L-myc fusion (204243_at), score: -0.48 RLN1relaxin 1 (211753_s_at), score: -0.76 RMND5Brequired for meiotic nuclear division 5 homolog B (S. cerevisiae) (218262_at), score: 0.49 RORARAR-related orphan receptor A (210426_x_at), score: -0.5 RP11-138L21.1similar to cell recognition molecule CASPR3 (220436_at), score: -0.46 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.55 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.51 RPAINRPA interacting protein (216961_s_at), score: -0.48 RPGRretinitis pigmentosa GTPase regulator (207624_s_at), score: 0.53 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.52 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.81 RPL38ribosomal protein L38 (202028_s_at), score: -0.52 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.62 RPS20ribosomal protein S20 (216247_at), score: -0.55 RPS27Aribosomal protein S27a (217144_at), score: -0.45 RRAGBRas-related GTP binding B (205540_s_at), score: -0.46 RRP8ribosomal RNA processing 8, methyltransferase, homolog (yeast) (203171_s_at), score: 0.83 RUFY2RUN and FYVE domain containing 2 (219957_at), score: -0.74 RYR3ryanodine receptor 3 (206306_at), score: -0.58 S100A14S100 calcium binding protein A14 (218677_at), score: -0.49 SALL1sal-like 1 (Drosophila) (206893_at), score: -0.52 SASH3SAM and SH3 domain containing 3 (204923_at), score: -0.53 SATB1SATB homeobox 1 (203408_s_at), score: -0.65 SBF1SET binding factor 1 (39835_at), score: 0.48 SBNO1strawberry notch homolog 1 (Drosophila) (218737_at), score: -0.5 SCAMP5secretory carrier membrane protein 5 (212699_at), score: -0.54 SCDstearoyl-CoA desaturase (delta-9-desaturase) (200832_s_at), score: -0.51 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: -0.45 SEPT5septin 5 (209767_s_at), score: -0.79 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: -0.46 SERPINB8serpin peptidase inhibitor, clade B (ovalbumin), member 8 (206034_at), score: 0.48 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.5 SETD6SET domain containing 6 (219751_at), score: -0.57 SFRS15splicing factor, arginine/serine-rich 15 (222310_at), score: -0.56 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.64 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: -0.53 SIK2salt-inducible kinase 2 (213221_s_at), score: -0.46 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.55 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.47 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: -0.55 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: -0.48 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.46 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.59 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.51 SLC35C1solute carrier family 35, member C1 (218485_s_at), score: 0.45 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.45 SLC46A3solute carrier family 46, member 3 (214719_at), score: -0.57 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: -0.49 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (212947_at), score: -0.53 SLCO1C1solute carrier organic anion transporter family, member 1C1 (220460_at), score: 0.74 SMAD6SMAD family member 6 (207069_s_at), score: -0.46 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: -0.68 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.63 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.48 SORL1sortilin-related receptor, L(DLR class) A repeats-containing (203509_at), score: -0.62 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: -0.52 SOX12SRY (sex determining region Y)-box 12 (204432_at), score: -0.62 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.5 SPASTspastin (209748_at), score: -0.45 SPATA1spermatogenesis associated 1 (221057_at), score: -0.46 SPATA6spermatogenesis associated 6 (220298_s_at), score: -0.55 SPEF1sperm flagellar 1 (216119_s_at), score: -0.52 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: -0.62 SRGAP2SLIT-ROBO Rho GTPase activating protein 2 (213329_at), score: -0.54 SSX3synovial sarcoma, X breakpoint 3 (215881_x_at), score: -0.5 ST7suppression of tumorigenicity 7 (207871_s_at), score: -0.58 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.79 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: -0.56 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: -0.44 SYT5synaptotagmin V (206161_s_at), score: -0.62 TAOK2TAO kinase 2 (204986_s_at), score: 0.64 TAPBPLTAP binding protein-like (218747_s_at), score: -0.45 TAS2R14taste receptor, type 2, member 14 (221391_at), score: -0.75 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.48 TBPTATA box binding protein (203135_at), score: 0.5 TBX21T-box 21 (220684_at), score: -0.58 TBX5T-box 5 (207155_at), score: -0.52 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.45 TFAMtranscription factor A, mitochondrial (203176_s_at), score: 0.48 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.63 THNSL1threonine synthase-like 1 (S. cerevisiae) (219800_s_at), score: 0.51 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.53 TM4SF20transmembrane 4 L six family member 20 (220639_at), score: -0.52 TMC5transmembrane channel-like 5 (219580_s_at), score: -0.79 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.54 TMEM22transmembrane protein 22 (219569_s_at), score: 0.47 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.66 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: -0.49 TNRC6Btrinucleotide repeat containing 6B (213254_at), score: -0.49 TOE1target of EGR1, member 1 (nuclear) (204080_at), score: 0.48 TRMT2BTRM2 tRNA methyltransferase 2 homolog B (S. cerevisiae) (205238_at), score: 0.51 TRNAU1APtRNA selenocysteine 1 associated protein 1 (218977_s_at), score: 0.7 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: 0.71 TSPAN9tetraspanin 9 (220968_s_at), score: -0.58 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: 0.54 TTBK2tau tubulin kinase 2 (213922_at), score: -0.48 TWISTNBTWIST neighbor (214729_at), score: 0.48 TXNthioredoxin (216609_at), score: -0.59 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: 0.59 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: -0.46 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.5 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: -0.57 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.8 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.73 VHLvon Hippel-Lindau tumor suppressor (203844_at), score: -0.73 WDR52WD repeat domain 52 (221103_s_at), score: 0.45 WDR60WD repeat domain 60 (219251_s_at), score: -0.47 WDR91WD repeat domain 91 (218971_s_at), score: -0.58 WFDC2WAP four-disulfide core domain 2 (203892_at), score: -0.67 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.66 XAF1XIAP associated factor 1 (206133_at), score: -0.47 XYLBxylulokinase homolog (H. influenzae) (214776_x_at), score: -0.72 ZBTB24zinc finger and BTB domain containing 24 (205340_at), score: -0.47 ZBTB7Azinc finger and BTB domain containing 7A (219186_at), score: -0.46 ZDHHC11zinc finger, DHHC-type containing 11 (221646_s_at), score: -0.66 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.45 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: -0.55 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.72 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.67 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.69 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.58 ZNF124zinc finger protein 124 (206928_at), score: -0.51 ZNF224zinc finger protein 224 (216983_s_at), score: -0.45 ZNF280Dzinc finger protein 280D (221213_s_at), score: -0.53 ZNF281zinc finger protein 281 (218401_s_at), score: -0.49 ZNF318zinc finger protein 318 (203521_s_at), score: 0.72 ZNF323zinc finger protein 323 (222016_s_at), score: 0.51 ZNF34zinc finger protein 34 (219801_at), score: -0.46 ZNF350zinc finger protein 350 (219266_at), score: -0.47 ZNF37Azinc finger protein 37A (214878_at), score: -0.69 ZNF516zinc finger protein 516 (203604_at), score: -0.5 ZNF668zinc finger protein 668 (219047_s_at), score: 0.47 ZNF93zinc finger protein 93 (208119_s_at), score: 0.47
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |