
Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cytokine binding
Interacting selectively with a cytokine, any of a group of proteins that function to control the survival, growth and differentiation of tissues and cells, and which have autocrine and paracrine activity.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
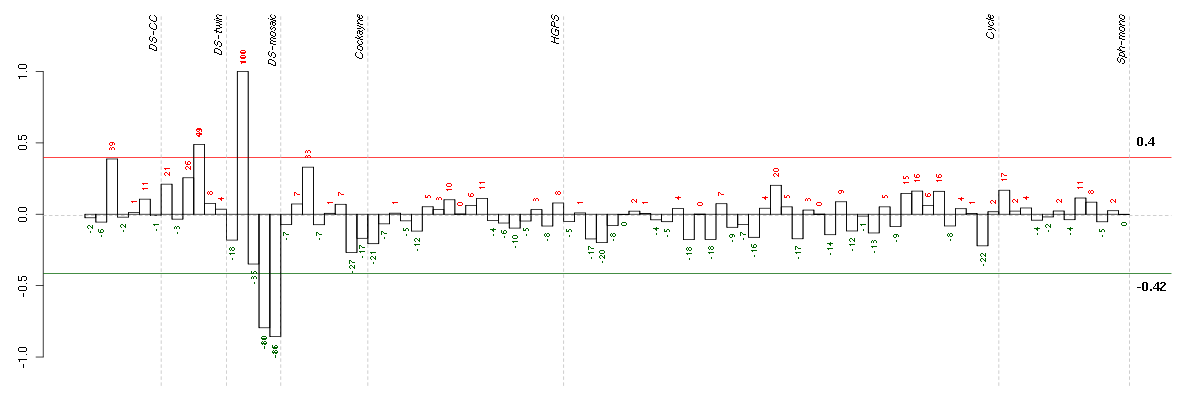
A2Malpha-2-macroglobulin (217757_at), score: 0.39 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: 0.49 ACOT8acyl-CoA thioesterase 8 (204212_at), score: -0.37 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: -0.3 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.6 ADAMTS2ADAM metallopeptidase with thrombospondin type 1 motif, 2 (214454_at), score: -0.47 ADAP2ArfGAP with dual PH domains 2 (219358_s_at), score: 0.53 ADCY7adenylate cyclase 7 (203741_s_at), score: 0.44 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.43 AEBP1AE binding protein 1 (201792_at), score: -0.32 AFF4AF4/FMR2 family, member 4 (219199_at), score: -0.33 AGFG1ArfGAP with FG repeats 1 (213926_s_at), score: -0.33 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.44 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: -0.34 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.43 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.47 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: -0.5 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.44 APLP2amyloid beta (A4) precursor-like protein 2 (208702_x_at), score: -0.32 APOC1apolipoprotein C-I (213553_x_at), score: 0.52 APOL1apolipoprotein L, 1 (209546_s_at), score: 0.42 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.44 APOL6apolipoprotein L, 6 (219716_at), score: 0.55 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: 0.43 ARF1ADP-ribosylation factor 1 (208750_s_at), score: -0.34 ARFRP1ADP-ribosylation factor related protein 1 (215984_s_at), score: -0.41 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: 0.58 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.64 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (209539_at), score: 0.41 ARPC4actin related protein 2/3 complex, subunit 4, 20kDa (211672_s_at), score: -0.54 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.45 ASPHaspartate beta-hydroxylase (205808_at), score: -0.32 ATP11AATPase, class VI, type 11A (215842_s_at), score: -0.33 ATP13A3ATPase type 13A3 (219558_at), score: -0.38 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.37 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: -0.35 AVILadvillin (205539_at), score: 0.46 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: -0.29 B9D1B9 protein domain 1 (210534_s_at), score: 0.62 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: -0.38 BAT2D1BAT2 domain containing 1 (211947_s_at), score: -0.54 BAXBCL2-associated X protein (208478_s_at), score: -0.34 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.42 BCHEbutyrylcholinesterase (205433_at), score: -0.75 BCL2L1BCL2-like 1 (215037_s_at), score: -0.54 BDKRB1bradykinin receptor B1 (207510_at), score: 0.48 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: -0.53 BHLHE41basic helix-loop-helix family, member e41 (221530_s_at), score: 0.39 BNC2basonuclin 2 (220272_at), score: 0.41 BRD2bromodomain containing 2 (214911_s_at), score: -0.37 BSGbasigin (Ok blood group) (208677_s_at), score: -0.37 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.32 BTRCbeta-transducin repeat containing (216091_s_at), score: -0.44 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.44 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.42 C15orf39chromosome 15 open reading frame 39 (204495_s_at), score: 0.4 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.37 C19orf29chromosome 19 open reading frame 29 (215954_s_at), score: -0.31 C19orf54chromosome 19 open reading frame 54 (222052_at), score: 0.5 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: -0.41 C1orf144chromosome 1 open reading frame 144 (212003_at), score: -0.36 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.4 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: -0.32 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: -0.62 C9orf91chromosome 9 open reading frame 91 (221865_at), score: 0.46 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: -0.36 CAMTA2calmodulin binding transcription activator 2 (212948_at), score: 0.44 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: 0.53 CASP9caspase 9, apoptosis-related cysteine peptidase (203984_s_at), score: 0.4 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: -0.31 CCND1cyclin D1 (208711_s_at), score: -0.46 CCScopper chaperone for superoxide dismutase (203522_at), score: 0.47 CD226CD226 molecule (207315_at), score: -0.35 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.46 CD82CD82 molecule (203904_x_at), score: -0.33 CDCP1CUB domain containing protein 1 (218451_at), score: 0.39 CDK10cyclin-dependent kinase 10 (210622_x_at), score: -0.44 CDK6cyclin-dependent kinase 6 (207143_at), score: -0.46 CDV3CDV3 homolog (mouse) (213548_s_at), score: -0.4 CEBPBCCAAT/enhancer binding protein (C/EBP), beta (212501_at), score: 0.46 CEBPDCCAAT/enhancer binding protein (C/EBP), delta (203973_s_at), score: 0.4 CECR5cat eye syndrome chromosome region, candidate 5 (218592_s_at), score: 0.4 CEP170centrosomal protein 170kDa (212746_s_at), score: -0.31 CFIcomplement factor I (203854_at), score: 0.79 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.51 CHKAcholine kinase alpha (204233_s_at), score: -0.31 CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (205567_at), score: 0.49 CHST11carbohydrate (chondroitin 4) sulfotransferase 11 (219634_at), score: -0.46 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: 0.47 CLK1CDC-like kinase 1 (214683_s_at), score: 0.43 CLK4CDC-like kinase 4 (210346_s_at), score: 0.42 CLPTM1cleft lip and palate associated transmembrane protein 1 (211136_s_at), score: -0.41 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.6 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.56 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.37 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.42 COL1A1collagen, type I, alpha 1 (217430_x_at), score: -0.58 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.4 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: -0.54 CPDcarboxypeptidase D (201942_s_at), score: -0.3 CPNE3copine III (202118_s_at), score: -0.43 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: -0.32 CRLF1cytokine receptor-like factor 1 (206315_at), score: -0.39 CTHcystathionase (cystathionine gamma-lyase) (217127_at), score: 0.48 CTSHcathepsin H (202295_s_at), score: 0.52 CTSL2cathepsin L2 (210074_at), score: 0.44 CUGBP1CUG triplet repeat, RNA binding protein 1 (204113_at), score: -0.34 CXorf40Bchromosome X open reading frame 40B (212961_x_at), score: -0.32 CYR61cysteine-rich, angiogenic inducer, 61 (210764_s_at), score: -0.35 CYTH3cytohesin 3 (206523_at), score: -0.47 CYTIPcytohesin 1 interacting protein (209606_at), score: -0.8 DAPK3death-associated protein kinase 3 (203890_s_at), score: -0.55 DCTN5dynactin 5 (p25) (209231_s_at), score: -0.43 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (212855_at), score: 0.41 DDA1DET1 and DDB1 associated 1 (218260_at), score: -0.4 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (200702_s_at), score: -0.45 DENND1ADENN/MADD domain containing 1A (219763_at), score: 0.42 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.58 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.38 DHRS11dehydrogenase/reductase (SDR family) member 11 (218756_s_at), score: 0.46 DHRS12dehydrogenase/reductase (SDR family) member 12 (204800_s_at), score: 0.39 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.56 DKFZp434H1419hypothetical protein DKFZp434H1419 (214717_at), score: -0.43 DKFZP586H2123regeneration associated muscle protease (213661_at), score: 0.43 DKK1dickkopf homolog 1 (Xenopus laevis) (204602_at), score: -0.52 DLGAP4discs, large (Drosophila) homolog-associated protein 4 (202570_s_at), score: -0.34 DNMBPdynamin binding protein (212838_at), score: -0.3 DOM3Zdom-3 homolog Z (C. elegans) (215982_s_at), score: 0.47 DOPEY2dopey family member 2 (205248_at), score: -0.34 DPM2dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit (209391_at), score: -0.3 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: -0.31 DSC2desmocollin 2 (204751_x_at), score: -0.59 DSC3desmocollin 3 (206032_at), score: -0.53 DSG2desmoglein 2 (217901_at), score: -0.32 DSPdesmoplakin (200606_at), score: -0.32 DSTdystonin (212253_x_at), score: -0.32 E2F5E2F transcription factor 5, p130-binding (221586_s_at), score: 0.46 ECHDC2enoyl Coenzyme A hydratase domain containing 2 (218552_at), score: 0.61 ECSITECSIT homolog (Drosophila) (218225_at), score: 0.48 EDC3enhancer of mRNA decapping 3 homolog (S. cerevisiae) (219207_at), score: -0.35 EDN1endothelin 1 (218995_s_at), score: -0.58 EFHC1EF-hand domain (C-terminal) containing 1 (219833_s_at), score: 0.6 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: 0.68 EHD1EH-domain containing 1 (209037_s_at), score: -0.29 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: -0.35 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.5 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: -0.53 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: -0.5 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.52 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: 0.63 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.35 ENC1ectodermal-neural cortex (with BTB-like domain) (201340_s_at), score: -0.32 ENSAendosulfine alpha (221487_s_at), score: -0.4 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: -0.35 ERC1ELKS/RAB6-interacting/CAST family member 1 (215606_s_at), score: -0.32 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: -0.3 ERLIN1ER lipid raft associated 1 (202444_s_at), score: -0.3 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: -0.34 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.34 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: -0.32 EVI1ecotropic viral integration site 1 (221884_at), score: -0.33 EZRezrin (208621_s_at), score: -0.51 FAM131Bfamily with sequence similarity 131, member B (205368_at), score: 0.4 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.42 FAM53Bfamily with sequence similarity 53, member B (203206_at), score: 0.43 FANCCFanconi anemia, complementation group C (205189_s_at), score: 0.49 FASNfatty acid synthase (212218_s_at), score: -0.31 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: -0.3 FBN1fibrillin 1 (202765_s_at), score: -0.37 FBXL18F-box and leucine-rich repeat protein 18 (215068_s_at), score: -0.32 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (203562_at), score: 0.55 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: -0.56 FGFR3fibroblast growth factor receptor 3 (204379_s_at), score: 0.48 FKRPfukutin related protein (219853_at), score: -0.42 FLJ10404hypothetical protein FLJ10404 (218920_at), score: 0.45 FLJ21075hypothetical protein FLJ21075 (221172_at), score: -0.31 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.4 FLOT2flotillin 2 (211299_s_at), score: -0.39 FN1fibronectin 1 (214701_s_at), score: -0.63 FNBP1formin binding protein 1 (212288_at), score: 0.45 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.37 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: 0.58 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.39 FRMD4AFERM domain containing 4A (208476_s_at), score: -0.43 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (210933_s_at), score: -0.41 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.3 GABPAGA binding protein transcription factor, alpha subunit 60kDa (210188_at), score: -0.34 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.5 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (201723_s_at), score: -0.31 GALNT10UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (207357_s_at), score: -0.42 GALNT2UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) (217787_s_at), score: -0.31 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.46 GCAgrancalcin, EF-hand calcium binding protein (203765_at), score: 0.41 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.71 GDF15growth differentiation factor 15 (221577_x_at), score: 0.41 GGA2golgi associated, gamma adaptin ear containing, ARF binding protein 2 (210658_s_at), score: -0.31 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.6 GLB1Lgalactosidase, beta 1-like (206540_at), score: 0.42 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: -0.41 GNEglucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase (205042_at), score: 0.4 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.45 GOLSYNGolgi-localized protein (218692_at), score: 0.5 GPC4glypican 4 (204983_s_at), score: 0.66 GPR137G protein-coupled receptor 137 (43934_at), score: -0.38 GPR175G protein-coupled receptor 175 (218855_at), score: -0.3 GPR176G protein-coupled receptor 176 (206673_at), score: -0.36 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.4 GPR56G protein-coupled receptor 56 (212070_at), score: 0.55 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.59 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.3 GSTZ1glutathione transferase zeta 1 (209531_at), score: 0.4 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.42 GTPBP2GTP binding protein 2 (221050_s_at), score: 0.41 HADHAhydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit (208629_s_at), score: -0.35 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: -0.54 HDAC9histone deacetylase 9 (205659_at), score: -0.34 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: 0.56 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.45 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.43 HIST1H2BDhistone cluster 1, H2bd (209911_x_at), score: -0.3 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: -0.43 HIST2H2BEhistone cluster 2, H2be (202708_s_at), score: -0.38 HIST3H2Ahistone cluster 3, H2a (221582_at), score: 0.46 HLXH2.0-like homeobox (214438_at), score: -0.36 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.47 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.47 HOXB9homeobox B9 (216417_x_at), score: -0.74 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: -0.45 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: -0.46 HS1BP3HCLS1 binding protein 3 (219020_at), score: 0.45 HSD11B1hydroxysteroid (11-beta) dehydrogenase 1 (205404_at), score: 0.42 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: -0.36 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: -0.33 IFNGR2interferon gamma receptor 2 (interferon gamma transducer 1) (201642_at), score: -0.4 IFT88intraflagellar transport 88 homolog (Chlamydomonas) (204703_at), score: 0.42 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.36 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: -0.32 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: 0.49 IL17RCinterleukin 17 receptor C (64440_at), score: -0.32 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.41 IL21Rinterleukin 21 receptor (219971_at), score: -0.38 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (211000_s_at), score: -0.3 ILF3interleukin enhancer binding factor 3, 90kDa (208930_s_at), score: -0.42 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.39 INHBEinhibin, beta E (210587_at), score: 1 IQGAP1IQ motif containing GTPase activating protein 1 (213446_s_at), score: -0.4 IRF1interferon regulatory factor 1 (202531_at), score: 0.58 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: 0.44 ITSN1intersectin 1 (SH3 domain protein) (209298_s_at), score: -0.31 JAM3junctional adhesion molecule 3 (212813_at), score: -0.39 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.52 JMJD2Bjumonji domain containing 2B (212492_s_at), score: -0.37 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: 0.43 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.48 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: -0.38 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: -0.51 KCTD5potassium channel tetramerisation domain containing 5 (218474_s_at), score: -0.37 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: -0.39 KIAA0100KIAA0100 (201729_s_at), score: -0.37 KIAA0556KIAA0556 (213185_at), score: 0.45 KIAA0644KIAA0644 gene product (205150_s_at), score: 0.51 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: -0.34 L1CAML1 cell adhesion molecule (204584_at), score: 0.53 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: -0.61 LARP5La ribonucleoprotein domain family, member 5 (208953_at), score: -0.39 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (219813_at), score: -0.38 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (218523_at), score: 0.67 LIFRleukemia inhibitory factor receptor alpha (205876_at), score: 0.4 LOC100128414similar to fibrinogen silencer binding protein (220549_at), score: -0.32 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.39 LOC285412similar to KIAA0737 protein (217448_s_at), score: -0.36 LOC391132similar to hCG2041276 (216177_at), score: 0.44 LOC646808hypothetical LOC646808 (222338_x_at), score: 0.46 LOC727977hypothetical LOC727977 (205787_x_at), score: -0.3 LOC729034similar to puromycin sensitive aminopeptidase (214107_x_at), score: -0.38 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.4 LOXlysyl oxidase (213640_s_at), score: -0.43 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.56 LRP12low density lipoprotein-related protein 12 (220253_s_at), score: -0.31 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.43 LSM12LSM12 homolog (S. cerevisiae) (212529_at), score: -0.35 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: -0.56 LUC7LLUC7-like (S. cerevisiae) (220143_x_at), score: 0.44 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.32 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.71 MADDMAP-kinase activating death domain (38398_at), score: 0.4 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: -0.35 MALLmal, T-cell differentiation protein-like (209373_at), score: 0.44 MAN2C1mannosidase, alpha, class 2C, member 1 (203668_at), score: 0.58 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: -0.45 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.48 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: -0.47 MAPRE3microtubule-associated protein, RP/EB family, member 3 (214270_s_at), score: -0.44 MARCH6membrane-associated ring finger (C3HC4) 6 (201737_s_at), score: -0.38 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: -0.38 MAXMYC associated factor X (210734_x_at), score: -0.56 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: -0.6 MCAMmelanoma cell adhesion molecule (210869_s_at), score: -0.37 MCFD2multiple coagulation factor deficiency 2 (212246_at), score: -0.3 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.55 MED31mediator complex subunit 31 (219318_x_at), score: -0.43 MEGF8multiple EGF-like-domains 8 (214778_at), score: -0.4 MFAP2microfibrillar-associated protein 2 (203417_at), score: -0.31 MFAP4microfibrillar-associated protein 4 (212713_at), score: 0.47 MFN2mitofusin 2 (216205_s_at), score: -0.6 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: 0.52 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: -0.35 MGMTO-6-methylguanine-DNA methyltransferase (204880_at), score: 0.39 MKNK1MAP kinase interacting serine/threonine kinase 1 (209467_s_at), score: 0.41 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.41 MLXMAX-like protein X (217909_s_at), score: -0.36 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: -0.54 MNTMAX binding protein (204206_at), score: 0.42 MOCOSmolybdenum cofactor sulfurase (219959_at), score: 0.61 MOCS3molybdenum cofactor synthesis 3 (206141_at), score: -0.37 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.39 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: -0.48 MPImannose phosphate isomerase (202472_at), score: 0.4 MRASmuscle RAS oncogene homolog (206538_at), score: -0.44 MRPS18Amitochondrial ribosomal protein S18A (221693_s_at), score: -0.45 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.6 MTMR15myotubularin related protein 15 (203678_at), score: 0.42 MTMR3myotubularin related protein 3 (202197_at), score: 0.43 MVDmevalonate (diphospho) decarboxylase (203027_s_at), score: -0.37 MVKmevalonate kinase (36907_at), score: 0.41 MYO1Dmyosin ID (212338_at), score: 0.43 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: -0.5 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.45 NAPAN-ethylmaleimide-sensitive factor attachment protein, alpha (206491_s_at), score: -0.45 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: -0.43 NCAM2neural cell adhesion molecule 2 (205669_at), score: 0.46 NCOA1nuclear receptor coactivator 1 (209106_at), score: 0.39 NCRNA00153non-protein coding RNA 153 (219961_s_at), score: 0.41 NDRG3NDRG family member 3 (221082_s_at), score: -0.31 NF2neurofibromin 2 (merlin) (211017_s_at), score: -0.32 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: -0.32 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.4 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: -0.33 NFKB2nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (207535_s_at), score: -0.37 NFYAnuclear transcription factor Y, alpha (204108_at), score: -0.43 NOS1nitric oxide synthase 1 (neuronal) (207309_at), score: 0.48 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: 0.44 NPnucleoside phosphorylase (201695_s_at), score: -0.29 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.43 NPTX1neuronal pentraxin I (204684_at), score: -0.41 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.63 NR2C1nuclear receptor subfamily 2, group C, member 1 (204791_at), score: 0.41 NRBP1nuclear receptor binding protein 1 (217765_at), score: -0.36 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: -0.55 OAZ2ornithine decarboxylase antizyme 2 (201364_s_at), score: -0.46 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.5 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.48 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.34 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: 0.61 OR7E38Polfactory receptor, family 7, subfamily E, member 38 pseudogene (217499_x_at), score: 0.44 OXTRoxytocin receptor (206825_at), score: -0.3 PACS1phosphofurin acidic cluster sorting protein 1 (220557_s_at), score: -0.54 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: -0.39 PAPPA2pappalysin 2 (213332_at), score: 0.68 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: 0.4 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: 0.48 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.54 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.54 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: -0.36 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: -0.51 PGS1phosphatidylglycerophosphate synthase 1 (219394_at), score: 0.4 PHACTR2phosphatase and actin regulator 2 (204048_s_at), score: -0.3 PIONpigeon homolog (Drosophila) (222150_s_at), score: 0.5 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: -0.46 PLAC8placenta-specific 8 (219014_at), score: -0.32 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: -0.45 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (201411_s_at), score: -0.45 PLS1plastin 1 (I isoform) (205190_at), score: -0.31 PMLpromyelocytic leukemia (206503_x_at), score: -0.3 PODNL1podocan-like 1 (220411_x_at), score: 0.43 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.49 POLIpolymerase (DNA directed) iota (219317_at), score: 0.4 POLR2Epolymerase (RNA) II (DNA directed) polypeptide E, 25kDa (213887_s_at), score: -0.3 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.36 POMT1protein-O-mannosyltransferase 1 (218476_at), score: 0.62 PPIFpeptidylprolyl isomerase F (201490_s_at), score: -0.32 PPP2R1Aprotein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform (200695_at), score: -0.41 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: -0.58 PPP5Cprotein phosphatase 5, catalytic subunit (201979_s_at), score: -0.33 PQLC1PQ loop repeat containing 1 (218208_at), score: 0.44 PRKYprotein kinase, Y-linked (206279_at), score: 0.45 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: -0.49 PRRX1paired related homeobox 1 (205991_s_at), score: -0.35 PSENENpresenilin enhancer 2 homolog (C. elegans) (218302_at), score: -0.3 PSG3pregnancy specific beta-1-glycoprotein 3 (215821_x_at), score: -0.3 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: -0.4 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: -0.42 PSMB10proteasome (prosome, macropain) subunit, beta type, 10 (202659_at), score: 0.43 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: -0.46 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: -0.6 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.51 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: 0.59 PTH1Rparathyroid hormone 1 receptor (205911_at), score: 0.45 PTMSparathymosin (218045_x_at), score: -0.51 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.3 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: -0.44 PTPN2protein tyrosine phosphatase, non-receptor type 2 (213136_at), score: 0.47 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: -0.6 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: 0.6 PTPRNprotein tyrosine phosphatase, receptor type, N (204945_at), score: 0.46 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: -0.45 PTRFpolymerase I and transcript release factor (208790_s_at), score: -0.58 PXNpaxillin (211823_s_at), score: -0.64 QKIquaking homolog, KH domain RNA binding (mouse) (214543_x_at), score: -0.31 RAB11BRAB11B, member RAS oncogene family (34478_at), score: -0.42 RAB3BRAB3B, member RAS oncogene family (205924_at), score: -0.35 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: -0.32 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: -0.36 RANGAP1Ran GTPase activating protein 1 (212125_at), score: -0.44 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: 0.78 RASA3RAS p21 protein activator 3 (206220_s_at), score: -0.58 RASA4RAS p21 protein activator 4 (208534_s_at), score: -0.5 RBM6RNA binding motif protein 6 (201967_at), score: 0.4 RCAN1regulator of calcineurin 1 (215253_s_at), score: -0.37 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.6 RGL2ral guanine nucleotide dissociation stimulator-like 2 (209110_s_at), score: 0.45 RGS17regulator of G-protein signaling 17 (220334_at), score: 0.53 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.42 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (212479_s_at), score: 0.42 RNF126ring finger protein 126 (205748_s_at), score: -0.34 RNF44ring finger protein 44 (203286_at), score: 0.42 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (219550_at), score: 0.54 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.46 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.32 RXRBretinoid X receptor, beta (215099_s_at), score: -0.53 SAMD14sterile alpha motif domain containing 14 (213866_at), score: -0.29 SCAPERS phase cyclin A-associated protein in the ER (209741_x_at), score: 0.4 SCMH1sex comb on midleg homolog 1 (Drosophila) (221216_s_at), score: 0.39 SCRG1scrapie responsive protein 1 (205475_at), score: 0.46 SEC14L1SEC14-like 1 (S. cerevisiae) (202082_s_at), score: -0.57 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: 0.44 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (205405_at), score: -0.43 SENP2SUMO1/sentrin/SMT3 specific peptidase 2 (218122_s_at), score: -0.3 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.37 SEPT11septin 11 (201308_s_at), score: -0.35 SEPT8septin 8 (209000_s_at), score: -0.31 SEPW1selenoprotein W, 1 (201194_at), score: -0.35 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.45 SERPINE1serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (202627_s_at), score: -0.33 SERPINI1serpin peptidase inhibitor, clade I (neuroserpin), member 1 (205352_at), score: -0.37 SETD8SET domain containing (lysine methyltransferase) 8 (220200_s_at), score: -0.38 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.32 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.44 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: -0.46 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: -0.45 SIRT5sirtuin (silent mating type information regulation 2 homolog) 5 (S. cerevisiae) (219185_at), score: 0.45 SKIv-ski sarcoma viral oncogene homolog (avian) (204270_at), score: -0.33 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: -0.47 SLC16A3solute carrier family 16, member 3 (monocarboxylic acid transporter 4) (202855_s_at), score: -0.29 SLC26A6solute carrier family 26, member 6 (221572_s_at), score: 0.74 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.64 SLC35E1solute carrier family 35, member E1 (222263_at), score: -0.45 SLC37A4solute carrier family 37 (glucose-6-phosphate transporter), member 4 (217289_s_at), score: -0.37 SLC39A9solute carrier family 39 (zinc transporter), member 9 (217859_s_at), score: -0.32 SLC46A3solute carrier family 46, member 3 (214719_at), score: -0.36 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: -0.41 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: -0.5 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (204588_s_at), score: 0.45 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: 0.39 SLCO2A1solute carrier organic anion transporter family, member 2A1 (204368_at), score: 0.39 SMARCA2SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (212257_s_at), score: -0.36 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.54 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.5 SNNstannin (218032_at), score: 0.4 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.33 SONSON DNA binding protein (201085_s_at), score: -0.45 SPIN1spindlin 1 (217813_s_at), score: -0.39 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: -0.55 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: 0.46 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: -0.43 SRPXsushi-repeat-containing protein, X-linked (204955_at), score: -0.32 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (221551_x_at), score: -0.38 STARD13StAR-related lipid transfer (START) domain containing 13 (213103_at), score: -0.44 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.39 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: -0.7 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: -0.3 STK32Bserine/threonine kinase 32B (219686_at), score: -0.43 SULF1sulfatase 1 (212353_at), score: 0.43 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: 0.62 SULT1A3sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 (210580_x_at), score: 0.54 SYT1synaptotagmin I (203999_at), score: 0.43 TAF4TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa (213090_s_at), score: 0.45 TANC2tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 (208425_s_at), score: -0.57 TAPBPLTAP binding protein-like (218747_s_at), score: 0.6 TBC1D12TBC1 domain family, member 12 (221858_at), score: 0.4 TBC1D3TBC1 domain family, member 3 (209403_at), score: 0.45 TBC1D5TBC1 domain family, member 5 (201813_s_at), score: -0.4 TBX3T-box 3 (219682_s_at), score: -0.3 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.51 TDRD3tudor domain containing 3 (208089_s_at), score: 0.48 TEKTEK tyrosine kinase, endothelial (206702_at), score: -0.43 TFEBtranscription factor EB (50221_at), score: 0.48 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: -0.5 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.49 THBS1thrombospondin 1 (201109_s_at), score: -0.36 THG1LtRNA-histidine guanylyltransferase 1-like (S. cerevisiae) (219122_s_at), score: 0.41 THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: 0.42 THY1Thy-1 cell surface antigen (208850_s_at), score: -0.35 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: -0.3 TLE1transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) (203221_at), score: 0.41 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: 0.4 TLR3toll-like receptor 3 (206271_at), score: 0.54 TM7SF2transmembrane 7 superfamily member 2 (210130_s_at), score: 0.46 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: -0.39 TMCO3transmembrane and coiled-coil domains 3 (220240_s_at), score: -0.4 TMEM109transmembrane protein 109 (201361_at), score: -0.39 TMEM161Atransmembrane protein 161A (218745_x_at), score: 0.51 TMEM204transmembrane protein 204 (219315_s_at), score: -0.32 TMEM48transmembrane protein 48 (218073_s_at), score: -0.32 TMOD3tropomodulin 3 (ubiquitous) (220800_s_at), score: -0.3 TNFAIP1tumor necrosis factor, alpha-induced protein 1 (endothelial) (201208_s_at), score: -0.31 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.49 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: -0.29 TNFRSF12Atumor necrosis factor receptor superfamily, member 12A (218368_s_at), score: -0.36 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: 0.47 TNFRSF1Btumor necrosis factor receptor superfamily, member 1B (203508_at), score: 0.42 TNFRSF6Btumor necrosis factor receptor superfamily, member 6b, decoy (206467_x_at), score: 0.43 TOLLIPtoll interacting protein (217930_s_at), score: -0.34 TOM1target of myb1 (chicken) (202807_s_at), score: 0.47 TOM1L1target of myb1 (chicken)-like 1 (204485_s_at), score: 0.46 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.61 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.5 TOP3Btopoisomerase (DNA) III beta (215781_s_at), score: -0.37 TOX4TOX high mobility group box family member 4 (201683_x_at), score: -0.42 TRAF1TNF receptor-associated factor 1 (205599_at), score: -0.53 TRAM2translocation associated membrane protein 2 (202368_s_at), score: -0.39 TRIB3tribbles homolog 3 (Drosophila) (218145_at), score: 0.4 TRIM2tripartite motif-containing 2 (202342_s_at), score: 0.46 TRIM25tripartite motif-containing 25 (206911_at), score: -0.32 TRIM3tripartite motif-containing 3 (204911_s_at), score: -0.4 TRIOtriple functional domain (PTPRF interacting) (208178_x_at), score: -0.42 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: 0.44 TRPV2transient receptor potential cation channel, subfamily V, member 2 (219282_s_at), score: -0.3 TSPAN13tetraspanin 13 (217979_at), score: -0.33 TSPYL2TSPY-like 2 (218012_at), score: 0.49 TTC3tetratricopeptide repeat domain 3 (208662_s_at), score: -0.32 TUBB2Atubulin, beta 2A (209372_x_at), score: -0.4 TWISTNBTWIST neighbor (214729_at), score: -0.34 TXNL4Bthioredoxin-like 4B (218794_s_at), score: 0.41 UBE2Mubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) (203109_at), score: -0.35 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.32 UBTFupstream binding transcription factor, RNA polymerase I (214881_s_at), score: -0.34 UCHL5IPUCHL5 interacting protein (213334_x_at), score: 0.44 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: -0.36 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.32 USP21ubiquitin specific peptidase 21 (218367_x_at), score: 0.4 USP3ubiquitin specific peptidase 3 (221654_s_at), score: 0.4 UTRNutrophin (213022_s_at), score: -0.39 VASPvasodilator-stimulated phosphoprotein (202205_at), score: -0.36 VAV2vav 2 guanine nucleotide exchange factor (205536_at), score: -0.4 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: 0.5 VEGFAvascular endothelial growth factor A (211527_x_at), score: -0.34 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: -0.36 WDR1WD repeat domain 1 (210935_s_at), score: -0.46 WDR68WD repeat domain 68 (221745_at), score: -0.41 WDR8WD repeat domain 8 (219322_s_at), score: 0.41 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: 0.43 XYLT1xylosyltransferase I (213725_x_at), score: -0.39 XYLT2xylosyltransferase II (219401_at), score: 0.39 YIPF5Yip1 domain family, member 5 (221423_s_at), score: -0.31 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: -0.34 ZCCHC11zinc finger, CCHC domain containing 11 (212704_at), score: 0.39 ZER1zer-1 homolog (C. elegans) (202456_s_at), score: -0.44 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: -0.47 ZFPL1zinc finger protein-like 1 (209428_s_at), score: -0.47 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: 0.42 ZNF134zinc finger protein 134 (206182_at), score: -0.3 ZNF467zinc finger protein 467 (214746_s_at), score: 0.7 ZNF688zinc finger protein 688 (213527_s_at), score: 0.65 ZNF711zinc finger protein 711 (207781_s_at), score: 0.4 ZNF783zinc finger family member 783 (221876_at), score: 0.39 ZSCAN5Azinc finger and SCAN domain containing 5A (219904_at), score: 0.4
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |