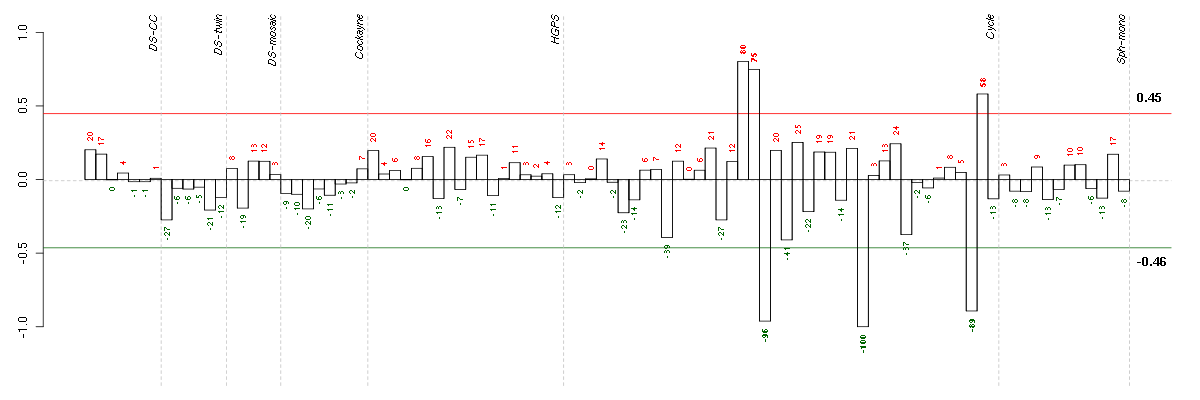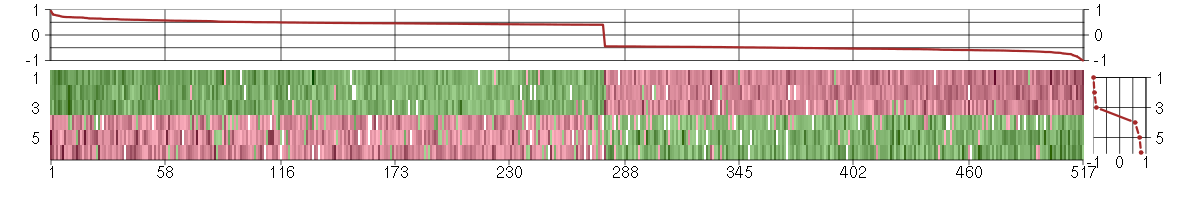

Under-expression is coded with green,
over-expression with red color.



ABL1c-abl oncogene 1, receptor tyrosine kinase (202123_s_at), score: 0.51 ACANaggrecan (205679_x_at), score: -0.52 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: -0.64 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.65 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.57 ADCY7adenylate cyclase 7 (203741_s_at), score: -0.49 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: -0.58 AGRNagrin (212285_s_at), score: 0.46 AJAP1adherens junctions associated protein 1 (206460_at), score: 0.41 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: 0.44 AMELXamelogenin (amelogenesis imperfecta 1, X-linked) (208410_x_at), score: -0.56 ANKRD28ankyrin repeat domain 28 (213035_at), score: -0.73 ANXA10annexin A10 (210143_at), score: 0.64 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: -0.6 AP3S2adaptor-related protein complex 3, sigma 2 subunit (202399_s_at), score: -0.51 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: -0.45 ARGLU1arginine and glutamate rich 1 (218067_s_at), score: 0.43 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.52 ARL17ADP-ribosylation factor-like 17 (210435_at), score: 0.48 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: -0.59 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: 0.4 ASTN2astrotactin 2 (209693_at), score: -0.52 ATAD5ATPase family, AAA domain containing 5 (220223_at), score: 0.45 ATXN2ataxin 2 (202622_s_at), score: -0.58 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.42 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: 0.49 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: -0.84 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.57 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: 0.5 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.48 BANK1B-cell scaffold protein with ankyrin repeats 1 (219667_s_at), score: -0.45 BAXBCL2-associated X protein (208478_s_at), score: -0.67 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: -0.59 BCL10B-cell CLL/lymphoma 10 (205263_at), score: 0.41 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (219528_s_at), score: 0.45 BCL2L2BCL2-like 2 (209311_at), score: 0.4 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: 0.42 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: -0.53 C10orf59chromosome 10 open reading frame 59 (220564_at), score: 0.42 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: -0.61 C12orf47chromosome 12 open reading frame 47 (64432_at), score: -0.47 C13orf27chromosome 13 open reading frame 27 (213346_at), score: 0.41 C16orf5chromosome 16 open reading frame 5 (218183_at), score: -0.53 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.47 C16orf7chromosome 16 open reading frame 7 (205781_at), score: 0.54 C17orf39chromosome 17 open reading frame 39 (220058_at), score: -0.54 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.48 C19orf54chromosome 19 open reading frame 54 (222052_at), score: -0.52 C1orf159chromosome 1 open reading frame 159 (219337_at), score: 0.56 C1orf50chromosome 1 open reading frame 50 (219406_at), score: -0.45 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: 0.49 C2CD2LC2CD2-like (204757_s_at), score: 0.5 C2orf49chromosome 2 open reading frame 49 (219662_at), score: -0.45 C3orf52chromosome 3 open reading frame 52 (219474_at), score: 0.44 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.41 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.53 C6orf48chromosome 6 open reading frame 48 (220755_s_at), score: -0.46 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: -0.59 C8orf17chromosome 8 open reading frame 17 (208266_at), score: 0.55 C8orf41chromosome 8 open reading frame 41 (219124_at), score: -0.45 C9orf114chromosome 9 open reading frame 114 (218565_at), score: -0.47 CA12carbonic anhydrase XII (203963_at), score: -0.48 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.43 CAMK2Bcalcium/calmodulin-dependent protein kinase II beta (34846_at), score: 0.51 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.49 CARD8caspase recruitment domain family, member 8 (204950_at), score: 0.46 CCDC94coiled-coil domain containing 94 (204335_at), score: -0.49 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.41 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: 0.44 CCNE1cyclin E1 (213523_at), score: 0.59 CD320CD320 molecule (218529_at), score: -0.74 CD40CD40 molecule, TNF receptor superfamily member 5 (35150_at), score: 0.45 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.51 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: 0.51 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.49 CDCA4cell division cycle associated 4 (218399_s_at), score: 0.69 CDCP1CUB domain containing protein 1 (218451_at), score: 0.48 CDH18cadherin 18, type 2 (206280_at), score: -0.59 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: -0.54 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: 0.45 CENPQcentromere protein Q (219294_at), score: 0.4 CENPTcentromere protein T (218148_at), score: 0.45 CEP70centrosomal protein 70kDa (219036_at), score: -0.55 CHPFchondroitin polymerizing factor (202175_at), score: 0.47 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: -0.53 CIRCBF1 interacting corepressor (209571_at), score: -0.46 CLCA2chloride channel accessory 2 (206165_s_at), score: 0.4 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.94 CLCN2chloride channel 2 (213499_at), score: -0.45 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.45 CLEC1AC-type lectin domain family 1, member A (219761_at), score: 0.48 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.68 CLIP4CAP-GLY domain containing linker protein family, member 4 (219944_at), score: 0.45 CLUclusterin (208791_at), score: 0.5 CNNM2cyclin M2 (206818_s_at), score: -0.6 CNNM3cyclin M3 (220739_s_at), score: 0.55 COASYCoenzyme A synthase (201913_s_at), score: -0.49 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.42 COL5A3collagen, type V, alpha 3 (52255_s_at), score: 0.79 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.69 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: -0.47 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: -0.56 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.53 CROCCL1ciliary rootlet coiled-coil, rootletin-like 1 (211038_s_at), score: -0.51 CRTC1CREB regulated transcription coactivator 1 (207159_x_at), score: 0.47 CRYL1crystallin, lambda 1 (220753_s_at), score: 0.49 CSNK2A2casein kinase 2, alpha prime polypeptide (203575_at), score: -0.46 CSRNP2cysteine-serine-rich nuclear protein 2 (221260_s_at), score: 0.42 CST6cystatin E/M (206595_at), score: 0.49 CTNScystinosis, nephropathic (36566_at), score: -0.49 CTSKcathepsin K (202450_s_at), score: 0.47 CTSOcathepsin O (203758_at), score: 0.51 CTSZcathepsin Z (210042_s_at), score: -0.63 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.45 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.61 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: 0.57 CYTH1cytohesin 1 (202880_s_at), score: 0.6 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: -0.52 DBC1deleted in bladder cancer 1 (205818_at), score: 0.42 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: -0.48 DEM1defects in morphology 1 homolog (S. cerevisiae) (219567_s_at), score: 0.46 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.7 DKFZp547G183hypothetical LOC55525 (220572_at), score: -0.54 DNAJC17DnaJ (Hsp40) homolog, subfamily C, member 17 (219861_at), score: -0.51 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.64 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.54 DNM3dynamin 3 (209839_at), score: 0.55 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: -0.56 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.51 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: -0.53 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.65 DUSP6dual specificity phosphatase 6 (208891_at), score: 0.76 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: -0.47 EDN1endothelin 1 (218995_s_at), score: 0.62 EDNRBendothelin receptor type B (204271_s_at), score: 0.71 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.48 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.55 EGR3early growth response 3 (206115_at), score: 0.6 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: 0.42 EIF3Heukaryotic translation initiation factor 3, subunit H (201592_at), score: -0.47 EIF3Leukaryotic translation initiation factor 3, subunit L (217719_at), score: -0.47 EIF4A2eukaryotic translation initiation factor 4A, isoform 2 (200912_s_at), score: -0.49 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: -0.46 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: 0.57 ELNelastin (212670_at), score: 0.45 EMCNendomucin (219436_s_at), score: -0.89 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.5 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.4 EPOerythropoietin (217254_s_at), score: -0.61 EREGepiregulin (205767_at), score: 0.55 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: 0.52 ESRRAestrogen-related receptor alpha (1487_at), score: 0.63 EVI2Aecotropic viral integration site 2A (204774_at), score: -0.58 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.63 EVLEnah/Vasp-like (217838_s_at), score: 0.47 EXOSC5exosome component 5 (218481_at), score: -0.57 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.43 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: -0.61 FAM131Afamily with sequence similarity 131, member A (221904_at), score: -0.55 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.55 FAM171A1family with sequence similarity 171, member A1 (212771_at), score: -0.54 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: 0.48 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: -0.62 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: 0.59 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: -0.47 FBXO31F-box protein 31 (219785_s_at), score: -0.66 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.54 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: 0.52 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: 0.61 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: 0.4 FKSG49FKSG49 (211454_x_at), score: -0.49 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.67 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.4 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.48 FOXJ2forkhead box J2 (203734_at), score: 0.48 FOXK2forkhead box K2 (203064_s_at), score: 0.49 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.48 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.7 GAAglucosidase, alpha; acid (202812_at), score: 0.69 GADD45Agrowth arrest and DNA-damage-inducible, alpha (203725_at), score: 0.46 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.51 GALNAC4S-6STB cell RAG associated protein (203066_at), score: 0.41 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: -0.54 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: -0.47 GANgigaxonin (220124_at), score: 0.49 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: -0.46 GDI1GDP dissociation inhibitor 1 (201864_at), score: 0.45 GIN1gypsy retrotransposon integrase 1 (219467_at), score: -0.46 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.53 GKglycerol kinase (207387_s_at), score: 0.48 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: -0.5 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: -0.65 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: -0.47 GPM6Bglycoprotein M6B (209167_at), score: 0.63 GPR137G protein-coupled receptor 137 (43934_at), score: 0.45 GPR153G protein-coupled receptor 153 (221902_at), score: 0.59 GPR172AG protein-coupled receptor 172A (222155_s_at), score: 0.43 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.44 GPR183G protein-coupled receptor 183 (205419_at), score: 0.47 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.56 GPSM3G-protein signaling modulator 3 (AGS3-like, C. elegans) (204265_s_at), score: -0.5 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.43 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: -0.59 HBBhemoglobin, beta (211696_x_at), score: 0.44 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.5 HERC3hect domain and RLD 3 (206183_s_at), score: 0.52 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.62 HINFPhistone H4 transcription factor (206495_s_at), score: -0.62 HISPPD2Ahistidine acid phosphatase domain containing 2A (204578_at), score: -0.46 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: -0.6 HIST1H2BEhistone cluster 1, H2be (208527_x_at), score: 0.43 HIST1H2BFhistone cluster 1, H2bf (208490_x_at), score: 0.47 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.56 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: 0.55 HLXH2.0-like homeobox (214438_at), score: 0.42 HNRNPA1heterogeneous nuclear ribonucleoprotein A1 (214280_x_at), score: -0.51 HNRNPA2B1heterogeneous nuclear ribonucleoprotein A2/B1 (205292_s_at), score: -0.46 HOXA11homeobox A11 (213823_at), score: -0.5 HOXD1homeobox D1 (205975_s_at), score: 0.45 HSPB2heat shock 27kDa protein 2 (205824_at), score: -0.58 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: -0.53 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.49 IER2immediate early response 2 (202081_at), score: 0.59 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.54 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.56 IGF2BP2insulin-like growth factor 2 mRNA binding protein 2 (218847_at), score: -0.51 IL11interleukin 11 (206924_at), score: 0.43 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: 0.56 IL33interleukin 33 (209821_at), score: 0.76 IMPA2inositol(myo)-1(or 4)-monophosphatase 2 (203126_at), score: 0.44 INHAinhibin, alpha (210141_s_at), score: -0.56 INTS8integrator complex subunit 8 (218905_at), score: -0.54 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.71 IRF6interferon regulatory factor 6 (202597_at), score: 0.42 ISOC2isochorismatase domain containing 2 (218893_at), score: -0.49 JMJD1Ajumonji domain containing 1A (212689_s_at), score: -0.53 JUNBjun B proto-oncogene (201473_at), score: 0.67 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: -0.63 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: 0.69 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.4 KCTD14potassium channel tetramerisation domain containing 14 (58916_at), score: -0.6 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: 0.46 KIAA0586KIAA0586 (205631_at), score: -0.63 KIF20Akinesin family member 20A (218755_at), score: -0.46 KIRRELkin of IRRE like (Drosophila) (220825_s_at), score: -0.56 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: -0.61 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: -0.51 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (205307_s_at), score: 0.41 KRT17keratin 17 (212236_x_at), score: 0.45 KRT33Akeratin 33A (208483_x_at), score: 0.58 LEMD3LEM domain containing 3 (218604_at), score: 0.49 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.69 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: -0.55 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: -0.79 LOC388152hypothetical LOC388152 (220602_s_at), score: -0.65 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.47 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: 0.46 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.46 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (219346_at), score: -0.48 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: 0.46 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.52 LRRC47leucine rich repeat containing 47 (212904_at), score: -0.47 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.48 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: 0.43 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: 0.56 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.8 MAP7D3MAP7 domain containing 3 (219576_at), score: -0.5 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.42 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.6 MED16mediator complex subunit 16 (43544_at), score: -0.53 MEF2Amyocyte enhancer factor 2A (214684_at), score: 0.51 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.4 MGAMAX gene associated (212945_s_at), score: -0.55 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.49 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.54 MLPHmelanophilin (218211_s_at), score: -0.61 MMACHCmethylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria (211774_s_at), score: -0.5 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: 0.5 MMP24matrix metallopeptidase 24 (membrane-inserted) (49679_s_at), score: 0.42 MMRN2multimerin 2 (219091_s_at), score: 0.41 MOSPD1motile sperm domain containing 1 (218853_s_at), score: -0.45 MSNmoesin (200600_at), score: 0.44 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: -0.67 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.47 MTMR3myotubularin related protein 3 (202197_at), score: 0.47 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.43 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: 0.64 MYH15myosin, heavy chain 15 (215331_at), score: -0.46 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: -0.46 NAGAN-acetylgalactosaminidase, alpha- (202943_s_at), score: -0.55 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: -0.5 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (219217_at), score: -0.47 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: 0.41 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.46 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.46 NEFLneurofilament, light polypeptide (221805_at), score: 0.58 NELFnasal embryonic LHRH factor (221214_s_at), score: 0.58 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: 0.73 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.47 NFICnuclear factor I/C (CCAAT-binding transcription factor) (206929_s_at), score: -0.49 NKX3-1NK3 homeobox 1 (209706_at), score: 0.44 NLKnemo-like kinase (218318_s_at), score: -0.45 NMBneuromedin B (205204_at), score: -0.6 NME6non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) (205851_at), score: -0.6 NOL8nucleolar protein 8 (218244_at), score: -0.6 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: 0.42 NR2F1nuclear receptor subfamily 2, group F, member 1 (209505_at), score: 0.46 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: 0.42 NSUN5BNOL1/NOP2/Sun domain family, member 5B (214100_x_at), score: -0.54 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: -0.53 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: -0.54 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: -0.54 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.56 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.46 OR7E38Polfactory receptor, family 7, subfamily E, member 38 pseudogene (217499_x_at), score: -0.45 OSBPoxysterol binding protein (201800_s_at), score: -0.47 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: -0.55 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: -0.6 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.46 PCDH9protocadherin 9 (219737_s_at), score: 0.42 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.43 PENKproenkephalin (213791_at), score: 0.51 PER2period homolog 2 (Drosophila) (205251_at), score: 0.44 PFDN6prefoldin subunit 6 (222029_x_at), score: -0.56 PFTK1PFTAIRE protein kinase 1 (204604_at), score: 0.46 PGPEP1pyroglutamyl-peptidase I (219891_at), score: -0.46 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: -0.45 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: -0.54 PIGCphosphatidylinositol glycan anchor biosynthesis, class C (216593_s_at), score: 0.43 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.61 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.42 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: -0.55 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.48 PKD2polycystic kidney disease 2 (autosomal dominant) (203688_at), score: 0.43 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: 0.49 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.58 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.6 PLP1proteolipid protein 1 (210198_s_at), score: -0.53 PLXNB1plexin B1 (215807_s_at), score: 0.59 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: 0.55 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (217610_at), score: -0.51 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.4 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.5 POMT1protein-O-mannosyltransferase 1 (218476_at), score: -0.54 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: -0.47 PORCNporcupine homolog (Drosophila) (219483_s_at), score: 0.47 PPFIBP1PTPRF interacting protein, binding protein 1 (liprin beta 1) (214374_s_at), score: -0.54 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: 0.45 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.42 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: 0.7 PRSS3protease, serine, 3 (213421_x_at), score: 0.61 PSD3pleckstrin and Sec7 domain containing 3 (203355_s_at), score: -0.49 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.54 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: -0.82 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.63 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.55 PTMAprothymosin, alpha (200772_x_at), score: -0.48 PTMAP7prothymosin, alpha pseudogene 7 (208549_x_at), score: -0.47 PXNpaxillin (211823_s_at), score: 0.41 QSOX1quiescin Q6 sulfhydryl oxidase 1 (201482_at), score: 0.48 R3HDM2R3H domain containing 2 (203831_at), score: -0.53 RAB36RAB36, member RAS oncogene family (211471_s_at), score: 0.4 RAB40BRAB40B, member RAS oncogene family (204547_at), score: 0.47 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: -0.47 RABL3RAB, member of RAS oncogene family-like 3 (213970_at), score: -0.7 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.47 RALGDSral guanine nucleotide dissociation stimulator (209050_s_at), score: -0.73 RASA2RAS p21 protein activator 2 (206636_at), score: 0.62 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.46 RBM39RNA binding motif protein 39 (207941_s_at), score: 0.4 RBM4BRNA binding motif protein 4B (209497_s_at), score: -0.51 RBMS3RNA binding motif, single stranded interacting protein (206767_at), score: -0.45 RCC1regulator of chromosome condensation 1 (206499_s_at), score: -0.59 RETret proto-oncogene (205879_x_at), score: 0.42 RFX7regulatory factor X, 7 (218430_s_at), score: -0.7 RHBDD3rhomboid domain containing 3 (204402_at), score: 0.52 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: -0.53 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.65 RIPK2receptor-interacting serine-threonine kinase 2 (209545_s_at), score: -0.46 RNF126ring finger protein 126 (205748_s_at), score: 0.42 RNF19Bring finger protein 19B (213038_at), score: 0.44 RNF2ring finger protein 2 (205215_at), score: 0.49 RNF44ring finger protein 44 (203286_at), score: -0.64 RPL14ribosomal protein L14 (219138_at), score: -0.47 RPL27Aribosomal protein L27a (212044_s_at), score: -0.45 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (204633_s_at), score: 0.47 S100A3S100 calcium binding protein A3 (206027_at), score: -0.46 SAMD9sterile alpha motif domain containing 9 (219691_at), score: 0.42 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: 0.65 SBF1SET binding factor 1 (39835_at), score: 0.44 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: 0.45 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.46 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.43 SETD6SET domain containing 6 (219751_at), score: -0.48 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: -0.95 SEZ6L2seizure related 6 homolog (mouse)-like 2 (218720_x_at), score: 0.56 SF1splicing factor 1 (208313_s_at), score: 0.5 SHROOM2shroom family member 2 (204967_at), score: 0.43 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.53 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.52 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: 0.6 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.63 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: 0.41 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: 0.6 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.57 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.5 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.75 SLC38A7solute carrier family 38, member 7 (218727_at), score: -0.45 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.4 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.46 SMAD6SMAD family member 6 (207069_s_at), score: -0.55 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.47 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: -0.55 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: -0.64 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.6 SNRKSNF related kinase (209481_at), score: 0.41 SNX13sorting nexin 13 (213292_s_at), score: -0.58 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.46 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.56 SPA17sperm autoantigenic protein 17 (205406_s_at), score: -0.59 SPAG1sperm associated antigen 1 (210117_at), score: 0.58 SPAG6sperm associated antigen 6 (210033_s_at), score: 0.5 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.56 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.63 SPTBN1spectrin, beta, non-erythrocytic 1 (212071_s_at), score: -0.49 SSFA2sperm specific antigen 2 (202506_at), score: 0.42 STC1stanniocalcin 1 (204597_x_at), score: 0.46 STK16serine/threonine kinase 16 (209622_at), score: -0.49 STX3syntaxin 3 (209238_at), score: -0.46 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: -0.52 SUV39H2suppressor of variegation 3-9 homolog 2 (Drosophila) (219262_at), score: 0.42 SYT1synaptotagmin I (203999_at), score: 0.52 SYT11synaptotagmin XI (209198_s_at), score: -0.69 TACC2transforming, acidic coiled-coil containing protein 2 (202289_s_at), score: -0.55 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.45 TAS2R9taste receptor, type 2, member 9 (221461_at), score: 0.46 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.44 TBPTATA box binding protein (203135_at), score: 0.43 TBX2T-box 2 (40560_at), score: 0.47 TBX5T-box 5 (207155_at), score: -0.46 TEAD3TEA domain family member 3 (209454_s_at), score: 0.41 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: -0.54 THNSL1threonine synthase-like 1 (S. cerevisiae) (219800_s_at), score: -0.45 THOC2THO complex 2 (212994_at), score: -0.59 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: -0.54 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.49 TIMM9translocase of inner mitochondrial membrane 9 homolog (yeast) (218316_at), score: -0.54 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: 0.5 TMEM158transmembrane protein 158 (213338_at), score: 0.47 TMEM160transmembrane protein 160 (219219_at), score: -0.68 TMEM22transmembrane protein 22 (219569_s_at), score: 0.44 TMSB15Athymosin beta 15a (205347_s_at), score: -0.66 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: 0.47 TNIKTRAF2 and NCK interacting kinase (213107_at), score: -0.67 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.47 TOP2Btopoisomerase (DNA) II beta 180kDa (211987_at), score: -0.46 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: -0.58 TP53TG1TP53 target 1 (non-protein coding) (209917_s_at), score: -0.46 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.7 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: -0.63 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: 0.5 TRIM14tripartite motif-containing 14 (203148_s_at), score: -0.46 TRIP11thyroid hormone receptor interactor 11 (209778_at), score: -0.6 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: -0.51 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: 0.44 TRPV4transient receptor potential cation channel, subfamily V, member 4 (219516_at), score: 0.52 TRY6trypsinogen C (215395_x_at), score: 0.44 TSKUtsukushin (218245_at), score: 0.42 TXNL4Bthioredoxin-like 4B (218794_s_at), score: -0.45 UBE3Aubiquitin protein ligase E3A (213291_s_at), score: 0.47 UCHL5IPUCHL5 interacting protein (213334_x_at), score: -0.53 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -1 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.48 USE1unconventional SNARE in the ER 1 homolog (S. cerevisiae) (221706_s_at), score: -0.57 USP36ubiquitin specific peptidase 36 (220370_s_at), score: 0.55 USP47ubiquitin specific peptidase 47 (221518_s_at), score: -0.51 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: -0.75 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.55 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.52 WDR3WD repeat domain 3 (218882_s_at), score: -0.49 WDR5BWD repeat domain 5B (219538_at), score: -0.5 WDR78WD repeat domain 78 (220769_s_at), score: 0.42 WIZwidely interspaced zinc finger motifs (52005_at), score: 0.4 XPO4exportin 4 (218479_s_at), score: -0.57 YIPF1Yip1 domain family, member 1 (214733_s_at), score: -0.5 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.5 YY2YY2 transcription factor (216531_at), score: -0.58 ZBTB24zinc finger and BTB domain containing 24 (205340_at), score: -0.59 ZC3H4zinc finger CCCH-type containing 4 (213390_at), score: -0.5 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.6 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: -0.48 ZKSCAN4zinc finger with KRAB and SCAN domains 4 (213625_at), score: -0.45 ZMAT5zinc finger, matrin type 5 (218752_at), score: -0.62 ZNF133zinc finger protein 133 (216960_s_at), score: -0.59 ZNF195zinc finger protein 195 (204234_s_at), score: -0.55 ZNF219zinc finger protein 219 (219314_s_at), score: 0.64 ZNF22zinc finger protein 22 (KOX 15) (218005_at), score: -0.67 ZNF239zinc finger protein 239 (206261_at), score: -0.48 ZNF248zinc finger protein 248 (213269_at), score: -0.54 ZNF277zinc finger protein 277 (218645_at), score: -0.45 ZNF280Dzinc finger protein 280D (221213_s_at), score: -0.57 ZNF324Bzinc finger protein 324B (222318_at), score: 0.46 ZNF337zinc finger protein 337 (37860_at), score: -0.46 ZNF350zinc finger protein 350 (219266_at), score: -0.5 ZNF434zinc finger protein 434 (218937_at), score: -0.48 ZNF480zinc finger protein 480 (222283_at), score: -0.62 ZNF510zinc finger protein 510 (206053_at), score: -0.46 ZNF536zinc finger protein 536 (206403_at), score: 0.41 ZNF654zinc finger protein 654 (219239_s_at), score: -0.46 ZNF667zinc finger protein 667 (207120_at), score: -0.62 ZNF688zinc finger protein 688 (213527_s_at), score: -0.48 ZNF749zinc finger protein 749 (215289_at), score: 0.5 ZNF93zinc finger protein 93 (208119_s_at), score: 0.41 ZRANB1zinc finger, RAN-binding domain containing 1 (201219_at), score: -0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |