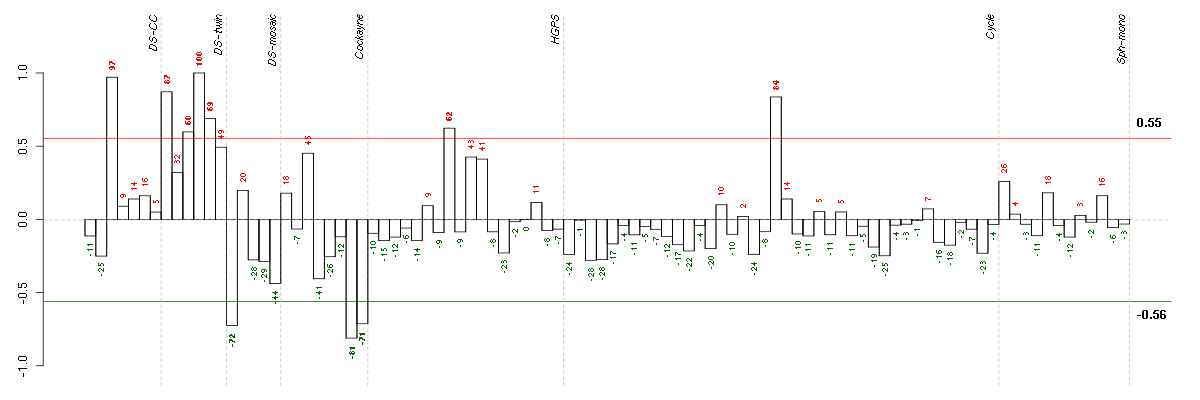

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell surface
The external part of the cell wall and/or plasma membrane.
endomembrane system
A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cytoskeletal protein binding
Interacting selectively with any protein component of any cytoskeleton (actin, microtubule, or intermediate filament cytoskeleton).
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-320/320abcd | 7.477e-03 | 22.53 | 47 | 372 |
ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.56 ACBD3acyl-Coenzyme A binding domain containing 3 (202323_s_at), score: -0.44 ACLYATP citrate lyase (210337_s_at), score: -0.62 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: -0.6 ADAM10ADAM metallopeptidase domain 10 (214895_s_at), score: -0.86 AFF4AF4/FMR2 family, member 4 (219199_at), score: -0.44 AGFG1ArfGAP with FG repeats 1 (213926_s_at), score: -0.63 AGPSalkylglycerone phosphate synthase (205401_at), score: -0.55 AHCYL1S-adenosylhomocysteine hydrolase-like 1 (200848_at), score: -0.41 AKIRIN1akirin 1 (217893_s_at), score: -0.48 ANGPT1angiopoietin 1 (205608_s_at), score: -0.37 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: -0.56 ANKLE2ankyrin repeat and LEM domain containing 2 (212201_at), score: -0.47 AP1S2adaptor-related protein complex 1, sigma 2 subunit (203300_x_at), score: -0.45 API5apoptosis inhibitor 5 (214959_s_at), score: -0.42 APLP2amyloid beta (A4) precursor-like protein 2 (208702_x_at), score: -0.47 ARF1ADP-ribosylation factor 1 (208750_s_at), score: -0.62 ARF6ADP-ribosylation factor 6 (203311_s_at), score: -0.47 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (216266_s_at), score: -0.77 ARIH1ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) (201881_s_at), score: -0.43 ARL2BPADP-ribosylation factor-like 2 binding protein (202092_s_at), score: -0.37 ARMC9armadillo repeat containing 9 (219637_at), score: -0.39 ARPP-19cyclic AMP phosphoprotein, 19 kD (214553_s_at), score: -0.48 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (221039_s_at), score: -0.55 ASPHaspartate beta-hydroxylase (205808_at), score: -0.75 ATP13A3ATPase type 13A3 (219558_at), score: -0.86 ATP6AP2ATPase, H+ transporting, lysosomal accessory protein 2 (201444_s_at), score: -0.46 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.4 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: -0.79 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.37 ATP6V1C1ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 (202874_s_at), score: -0.42 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: -0.4 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: -0.47 BAG5BCL2-associated athanogene 5 (202984_s_at), score: -0.59 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.46 BCAT1branched chain aminotransferase 1, cytosolic (214452_at), score: -0.43 BCHEbutyrylcholinesterase (205433_at), score: -0.58 BCL2L1BCL2-like 1 (215037_s_at), score: -0.48 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: -0.64 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: -0.44 BICD2bicaudal D homolog 2 (Drosophila) (209203_s_at), score: -0.58 BMPR2bone morphogenetic protein receptor, type II (serine/threonine kinase) (210214_s_at), score: -0.67 BSGbasigin (Ok blood group) (208677_s_at), score: -0.4 C14orf1chromosome 14 open reading frame 1 (217188_s_at), score: -0.64 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: -0.49 C18orf10chromosome 18 open reading frame 10 (213617_s_at), score: -0.5 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: -0.57 C1orf144chromosome 1 open reading frame 144 (212003_at), score: -0.43 C1QTNF1C1q and tumor necrosis factor related protein 1 (220975_s_at), score: 0.84 C20orf30chromosome 20 open reading frame 30 (220477_s_at), score: -0.73 C2CD2LC2CD2-like (204757_s_at), score: 0.93 C4orf18chromosome 4 open reading frame 18 (219872_at), score: -0.53 CALD1caldesmon 1 (201616_s_at), score: -0.57 CALUcalumenin (214845_s_at), score: -0.77 CANXcalnexin (208853_s_at), score: -0.61 CAPN7calpain 7 (203357_s_at), score: -0.65 CAPRIN1cell cycle associated protein 1 (200722_s_at), score: -0.72 CBFBcore-binding factor, beta subunit (206788_s_at), score: -0.76 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.53 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: -0.74 CDC27cell division cycle 27 homolog (S. cerevisiae) (217881_s_at), score: -0.47 CDH11cadherin 11, type 2, OB-cadherin (osteoblast) (207172_s_at), score: -0.5 CDV3CDV3 homolog (mouse) (213548_s_at), score: -0.97 CDYLchromodomain protein, Y-like (203100_s_at), score: -0.51 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: -0.45 CLDND1claudin domain containing 1 (208925_at), score: -0.39 CLIC4chloride intracellular channel 4 (201559_s_at), score: -0.76 CLINT1clathrin interactor 1 (201768_s_at), score: -0.54 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: -0.71 COL4A1collagen, type IV, alpha 1 (211981_at), score: -0.38 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.7 COMTcatechol-O-methyltransferase (208817_at), score: -0.38 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: -0.96 COPS8COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) (202143_s_at), score: -0.47 CPDcarboxypeptidase D (201942_s_at), score: -1 CPNE3copine III (202118_s_at), score: -0.53 CRKv-crk sarcoma virus CT10 oncogene homolog (avian) (202226_s_at), score: -0.41 CSF1colony stimulating factor 1 (macrophage) (209716_at), score: -0.51 CSNK1A1casein kinase 1, alpha 1 (206562_s_at), score: -0.49 CTTNcortactin (201059_at), score: 0.87 CUL4Bcullin 4B (202213_s_at), score: -0.51 CYP51A1cytochrome P450, family 51, subfamily A, polypeptide 1 (216607_s_at), score: -0.88 DAPK3death-associated protein kinase 3 (203890_s_at), score: -0.65 DAZAP2DAZ associated protein 2 (212595_s_at), score: -0.87 DCTN4dynactin 4 (p62) (218013_x_at), score: -0.43 DCTN5dynactin 5 (p25) (209231_s_at), score: -0.38 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.92 DEGS1degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) (207431_s_at), score: -0.49 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: 0.84 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.43 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.93 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (210802_s_at), score: -0.57 DLG1discs, large homolog 1 (Drosophila) (202514_at), score: -0.44 DNAJB1DnaJ (Hsp40) homolog, subfamily B, member 1 (200664_s_at), score: -0.7 DNAJB4DnaJ (Hsp40) homolog, subfamily B, member 4 (203811_s_at), score: -0.57 DNASE2deoxyribonuclease II, lysosomal (214992_s_at), score: -0.46 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: -0.4 DSTdystonin (212253_x_at), score: -0.55 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: -0.57 EDN1endothelin 1 (218995_s_at), score: -0.42 EGR3early growth response 3 (206115_at), score: -0.41 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: -0.53 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: -0.38 EML4echinoderm microtubule associated protein like 4 (220386_s_at), score: -0.54 ENSAendosulfine alpha (221487_s_at), score: -0.47 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: -0.61 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: -0.56 ERLIN1ER lipid raft associated 1 (202444_s_at), score: -0.76 EXOC5exocyst complex component 5 (218748_s_at), score: -0.82 EZRezrin (208621_s_at), score: -0.56 FADS3fatty acid desaturase 3 (216080_s_at), score: -0.39 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: -0.66 FBN1fibrillin 1 (202765_s_at), score: -0.5 FGF5fibroblast growth factor 5 (210310_s_at), score: -0.63 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.39 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: -0.38 FHL1four and a half LIM domains 1 (201539_s_at), score: -0.44 FHOD1formin homology 2 domain containing 1 (218530_at), score: -0.42 FKBP15FK506 binding protein 15, 133kDa (76897_s_at), score: -0.51 FKBP1AFK506 binding protein 1A, 12kDa (210186_s_at), score: -0.78 FN1fibronectin 1 (214701_s_at), score: -0.75 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.45 FUSIP1FUS interacting protein (serine/arginine-rich) 1 (210178_x_at), score: -0.45 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: -0.52 FZD6frizzled homolog 6 (Drosophila) (203987_at), score: -0.39 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: -0.45 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: -0.7 GDI2GDP dissociation inhibitor 2 (200008_s_at), score: -0.44 GLUD1glutamate dehydrogenase 1 (200946_x_at), score: -0.55 GLUD2glutamate dehydrogenase 2 (215794_x_at), score: -0.61 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: -0.4 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: -0.86 GNAI3guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 (201179_s_at), score: -0.46 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: 0.85 GNRHR2gonadotropin-releasing hormone (type 2) receptor 2 (213852_at), score: 0.86 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: -0.96 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.9 GPC4glypican 4 (204983_s_at), score: -0.44 GPR137G protein-coupled receptor 137 (43934_at), score: -0.55 GPR176G protein-coupled receptor 176 (206673_at), score: -0.6 GPR183G protein-coupled receptor 183 (205419_at), score: -0.43 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: -0.48 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.38 GSTM4glutathione S-transferase mu 4 (204149_s_at), score: -0.4 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.84 H2AFYH2A histone family, member Y (214500_at), score: -0.43 H2BFSH2B histone family, member S (208579_x_at), score: -0.37 HADHAhydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit (208629_s_at), score: -0.49 HERC4hect domain and RLD 4 (208055_s_at), score: -0.5 HHEXhematopoietically expressed homeobox (215933_s_at), score: -0.49 HIG2hypoxia-inducible protein 2 (218507_at), score: 0.94 HIP1huntingtin interacting protein 1 (205425_at), score: -0.53 HIP1Rhuntingtin interacting protein 1 related (209558_s_at), score: 0.83 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: -0.78 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.42 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: -0.55 HOXB5homeobox B5 (205601_s_at), score: -0.38 HOXB6homeobox B6 (205366_s_at), score: -0.68 HOXB7homeobox B7 (204779_s_at), score: -0.4 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: -0.64 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: -0.77 HSPH1heat shock 105kDa/110kDa protein 1 (208744_x_at), score: -0.45 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.38 IFNGR1interferon gamma receptor 1 (211676_s_at), score: -0.54 IL13RA1interleukin 13 receptor, alpha 1 (210904_s_at), score: -0.58 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: -0.44 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.5 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (211000_s_at), score: -0.63 IPO8importin 8 (205701_at), score: -0.57 ITGA1integrin, alpha 1 (214660_at), score: -0.38 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (205885_s_at), score: -0.77 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: -0.58 KDELR3KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 (207265_s_at), score: -0.37 KHDRBS3KH domain containing, RNA binding, signal transduction associated 3 (209781_s_at), score: -0.41 KIAA1033KIAA1033 (215936_s_at), score: -0.46 KIAA1467KIAA1467 (213234_at), score: 0.87 KIDINS220kinase D-interacting substrate, 220kDa (212162_at), score: -0.51 KIF5Bkinesin family member 5B (201992_s_at), score: -0.46 KIFC3kinesin family member C3 (209661_at), score: 0.9 KITLGKIT ligand (211124_s_at), score: -0.67 KLHL20kelch-like 20 (Drosophila) (210635_s_at), score: -0.44 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: -0.89 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: -0.53 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.37 LARP5La ribonucleoprotein domain family, member 5 (208953_at), score: -0.56 LDLRlow density lipoprotein receptor (202068_s_at), score: -0.41 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.63 LIMS1LIM and senescent cell antigen-like domains 1 (207198_s_at), score: -0.46 LIN7Clin-7 homolog C (C. elegans) (219399_at), score: -0.39 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: -0.85 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: -0.44 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: -0.37 LOC100128809similar to hCG2045829 (215707_s_at), score: -0.62 LOC100133166similar to neighbor of BRCA1 gene 1 (201383_s_at), score: -0.54 LOC285412similar to KIAA0737 protein (217448_s_at), score: -0.57 LOC643287similar to prothymosin alpha (216384_x_at), score: 0.87 LOC727977hypothetical LOC727977 (205787_x_at), score: -0.38 LOC729148nuclear undecaprenyl pyrophosphate synthase 1 pseudogene (215207_x_at), score: -0.54 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.51 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.42 LYPLA2P1lysophospholipase II pseudogene 1 (216606_x_at), score: 0.82 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.63 MADDMAP-kinase activating death domain (38398_at), score: 0.95 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.5 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: -0.55 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.94 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: -0.73 MAP3K7IP2mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (210284_s_at), score: -0.89 MAP4K5mitogen-activated protein kinase kinase kinase kinase 5 (203553_s_at), score: -0.67 MAPK1mitogen-activated protein kinase 1 (208351_s_at), score: -0.82 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: -0.78 MAPRE1microtubule-associated protein, RP/EB family, member 1 (200712_s_at), score: -0.43 MAPRE2microtubule-associated protein, RP/EB family, member 2 (202501_at), score: -0.39 MARCH7membrane-associated ring finger (C3HC4) 7 (202654_x_at), score: -0.77 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: -0.85 MAXMYC associated factor X (210734_x_at), score: -0.85 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: -0.48 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: -0.81 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: -0.42 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (210154_at), score: -0.48 MED6mediator complex subunit 6 (207079_s_at), score: -0.54 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: -0.44 MFN2mitofusin 2 (216205_s_at), score: -0.61 MGAT2mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (211061_s_at), score: -0.42 MOBKL1BMOB1, Mps One Binder kinase activator-like 1B (yeast) (201299_s_at), score: -0.59 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.91 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: -0.37 MR1major histocompatibility complex, class I-related (207565_s_at), score: -0.39 MTMR2myotubularin related protein 2 (214649_s_at), score: -0.5 NAB1NGFI-A binding protein 1 (EGR1 binding protein 1) (208047_s_at), score: -0.71 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: -0.48 NCK1NCK adaptor protein 1 (204725_s_at), score: -0.37 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.37 NEU1sialidase 1 (lysosomal sialidase) (208926_at), score: -0.39 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: -0.39 NFYBnuclear transcription factor Y, beta (218129_s_at), score: -0.39 NID1nidogen 1 (202008_s_at), score: -0.79 NMD3NMD3 homolog (S. cerevisiae) (218036_x_at), score: -0.54 NNTnicotinamide nucleotide transhydrogenase (202784_s_at), score: -0.6 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: -0.78 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: -0.5 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: -0.41 NUP50nucleoporin 50kDa (218295_s_at), score: -0.47 OSMRoncostatin M receptor (205729_at), score: -0.79 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: -0.79 PALLDpalladin, cytoskeletal associated protein (200906_s_at), score: -0.43 PAPOLApoly(A) polymerase alpha (212720_at), score: -0.69 PARP4poly (ADP-ribose) polymerase family, member 4 (202239_at), score: -0.49 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.96 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.8 PCYOX1prenylcysteine oxidase 1 (203803_at), score: -0.53 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.37 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.43 PDLIM5PDZ and LIM domain 5 (216804_s_at), score: -0.41 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: 0.84 PGRMC1progesterone receptor membrane component 1 (201120_s_at), score: -0.53 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: -0.93 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: -0.85 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: -0.56 PKP2plakophilin 2 (207717_s_at), score: -0.46 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.37 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.44 PON2paraoxonase 2 (210830_s_at), score: -0.49 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: -0.37 PREPLprolyl endopeptidase-like (212216_at), score: -0.41 PRKAA1protein kinase, AMP-activated, alpha 1 catalytic subunit (209799_at), score: -0.5 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: -0.66 PRKAR1Aprotein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) (200604_s_at), score: -0.63 PRKD3protein kinase D3 (211084_x_at), score: -0.55 PROX1prospero homeobox 1 (207401_at), score: 0.84 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: -0.41 PRPF4BPRP4 pre-mRNA processing factor 4 homolog B (yeast) (211090_s_at), score: -0.84 PRRX1paired related homeobox 1 (205991_s_at), score: -0.8 PSEN1presenilin 1 (207782_s_at), score: -0.7 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.37 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: -0.49 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: -0.77 PTMAP7prothymosin, alpha pseudogene 7 (208549_x_at), score: 0.84 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.41 PTPLAD1protein tyrosine phosphatase-like A domain containing 1 (217777_s_at), score: -0.41 PTPN11protein tyrosine phosphatase, non-receptor type 11 (209896_s_at), score: -0.58 PTPN12protein tyrosine phosphatase, non-receptor type 12 (216915_s_at), score: -0.64 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: -0.67 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: -0.6 PTPRNprotein tyrosine phosphatase, receptor type, N (204945_at), score: 0.85 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.57 PXNpaxillin (211823_s_at), score: -0.74 RAB35RAB35, member RAS oncogene family (205461_at), score: -0.47 RAB3GAP1RAB3 GTPase activating protein subunit 1 (catalytic) (212932_at), score: -0.46 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: -0.92 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: -0.78 RALBv-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein) (202101_s_at), score: -0.37 RANBP2RAN binding protein 2 (201711_x_at), score: -0.8 RANBP9RAN binding protein 9 (202583_s_at), score: -0.64 RASA3RAS p21 protein activator 3 (206220_s_at), score: -0.47 RBPJrecombination signal binding protein for immunoglobulin kappa J region (211974_x_at), score: 0.85 RCAN1regulator of calcineurin 1 (215253_s_at), score: -0.54 RCOR3REST corepressor 3 (218344_s_at), score: -0.41 RDXradixin (204969_s_at), score: -0.69 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.44 RGPD5RANBP2-like and GRIP domain containing 5 (210676_x_at), score: -0.54 RGS4regulator of G-protein signaling 4 (204338_s_at), score: -0.48 RIOK3RIO kinase 3 (yeast) (202129_s_at), score: -0.58 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.43 RNF13ring finger protein 13 (201779_s_at), score: -0.49 RNF141ring finger protein 141 (219104_at), score: -0.37 RNF187ring finger protein 187 (212155_at), score: 0.83 RNF6ring finger protein (C3H2C3 type) 6 (210932_s_at), score: -0.45 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.62 RNGTTRNA guanylyltransferase and 5'-phosphatase (204207_s_at), score: -0.42 RRADRas-related associated with diabetes (204802_at), score: -0.37 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: -0.42 RRP7Bribosomal RNA processing 7 homolog B (S. cerevisiae) (214828_s_at), score: -0.47 RXRBretinoid X receptor, beta (215099_s_at), score: -0.52 RYKRYK receptor-like tyrosine kinase (216976_s_at), score: -0.55 SASH3SAM and SH3 domain containing 3 (204923_at), score: 0.83 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: -0.78 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: -0.81 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: -0.49 SCP2sterol carrier protein 2 (201339_s_at), score: -0.37 SCYL2SCY1-like 2 (S. cerevisiae) (221220_s_at), score: -0.63 SDHCsuccinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa (216591_s_at), score: -0.59 SEC14L1SEC14-like 1 (S. cerevisiae) (202082_s_at), score: -0.38 SEC23ASec23 homolog A (S. cerevisiae) (204344_s_at), score: -0.47 SEC23IPSEC23 interacting protein (216392_s_at), score: -0.6 SEC24ASEC24 family, member A (S. cerevisiae) (212902_at), score: -0.48 SELTselenoprotein T (217811_at), score: -0.42 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: 0.84 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.59 SEPT10septin 10 (214720_x_at), score: -0.56 SEPT11septin 11 (201308_s_at), score: -0.65 SEPT2septin 2 (200778_s_at), score: -0.37 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.86 SFRS1splicing factor, arginine/serine-rich 1 (201742_x_at), score: -0.5 SFRS2Bsplicing factor, arginine/serine-rich 2B (213152_s_at), score: -0.52 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: -0.5 SGCBsarcoglycan, beta (43kDa dystrophin-associated glycoprotein) (205121_at), score: -0.56 SGPP1sphingosine-1-phosphate phosphatase 1 (221268_s_at), score: -0.75 SH3BGRLSH3 domain binding glutamic acid-rich protein like (201311_s_at), score: -0.46 SHPKsedoheptulokinase (219713_at), score: 0.9 SKAP2src kinase associated phosphoprotein 2 (216899_s_at), score: -0.72 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: -0.49 SLC39A9solute carrier family 39 (zinc transporter), member 9 (217859_s_at), score: -0.41 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (203578_s_at), score: -0.4 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.75 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.76 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (208608_s_at), score: -0.43 SNX4sorting nexin 4 (205329_s_at), score: -0.37 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: -0.38 SPAG9sperm associated antigen 9 (206748_s_at), score: -0.71 SPIN1spindlin 1 (217813_s_at), score: -0.43 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: -0.38 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: 0.87 SPTLC1serine palmitoyltransferase, long chain base subunit 1 (202278_s_at), score: -0.7 SQLEsqualene epoxidase (213562_s_at), score: -0.69 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: -0.62 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: -0.71 SSFA2sperm specific antigen 2 (202506_at), score: -0.38 STAT1signal transducer and activator of transcription 1, 91kDa (AFFX-HUMISGF3A/M97935_MA_at), score: -0.65 STAT3signal transducer and activator of transcription 3 (acute-phase response factor) (208992_s_at), score: -0.52 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: -0.7 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.38 STOMstomatin (201060_x_at), score: -0.64 SYPL1synaptophysin-like 1 (201259_s_at), score: -0.77 TAF12TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa (209463_s_at), score: -0.4 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: -0.76 TANKTRAF family member-associated NFKB activator (207616_s_at), score: -0.38 TBL1XR1transducin (beta)-like 1 X-linked receptor 1 (221428_s_at), score: -0.74 TBX3T-box 3 (219682_s_at), score: -0.42 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (34260_at), score: 0.83 TFCP2transcription factor CP2 (207627_s_at), score: -0.4 TFDP1transcription factor Dp-1 (204147_s_at), score: -0.5 TFPItissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) (209676_at), score: -0.39 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: -0.46 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.79 TJP1tight junction protein 1 (zona occludens 1) (214168_s_at), score: -0.72 TLK2tousled-like kinase 2 (212986_s_at), score: -0.39 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: -0.44 TM9SF1transmembrane 9 superfamily member 1 (209149_s_at), score: -0.41 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: -0.52 TMCO3transmembrane and coiled-coil domains 3 (220240_s_at), score: -0.44 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: -0.63 TMOD3tropomodulin 3 (ubiquitous) (220800_s_at), score: -0.48 TMX1thioredoxin-related transmembrane protein 1 (208097_s_at), score: -0.7 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.37 TNFRSF11Btumor necrosis factor receptor superfamily, member 11b (204933_s_at), score: -0.47 TNFRSF21tumor necrosis factor receptor superfamily, member 21 (214581_x_at), score: -0.4 TOR1AIP1torsin A interacting protein 1 (216100_s_at), score: -0.68 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: -0.53 TPM4tropomyosin 4 (212481_s_at), score: -0.4 TPP1tripeptidyl peptidase I (200742_s_at), score: -0.63 TRIM22tripartite motif-containing 22 (213293_s_at), score: -0.43 TSNtranslin (201504_s_at), score: -0.58 TSPAN3tetraspanin 3 (200973_s_at), score: -0.47 TTPALtocopherol (alpha) transfer protein-like (219633_at), score: -0.43 TUBB2Atubulin, beta 2A (209372_x_at), score: -0.39 TWF1twinfilin, actin-binding protein, homolog 1 (Drosophila) (214007_s_at), score: -0.81 TWSG1twisted gastrulation homolog 1 (Drosophila) (219201_s_at), score: -0.48 UBA6ubiquitin-like modifier activating enzyme 6 (218340_s_at), score: -0.39 UBE2D3ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) (200669_s_at), score: -0.54 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: -0.51 UBXN4UBX domain protein 4 (212008_at), score: -0.82 USO1USO1 homolog, vesicle docking protein (yeast) (201831_s_at), score: -0.44 USP24ubiquitin specific peptidase 24 (212381_at), score: -0.38 USP34ubiquitin specific peptidase 34 (212065_s_at), score: -0.63 UTRNutrophin (213022_s_at), score: -0.57 VAMP3vesicle-associated membrane protein 3 (cellubrevin) (201337_s_at), score: -0.78 VCLvinculin (200930_s_at), score: -0.45 VEGFAvascular endothelial growth factor A (211527_x_at), score: -0.59 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: -0.96 WDR1WD repeat domain 1 (210935_s_at), score: -0.62 WIPF1WAS/WASL interacting protein family, member 1 (202663_at), score: -0.44 WTAPWilms tumor 1 associated protein (210285_x_at), score: -0.43 XPNPEP1X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (208453_s_at), score: -0.49 YIPF5Yip1 domain family, member 5 (221423_s_at), score: -0.61 YWHAEtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (210317_s_at), score: -0.44 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: -0.65 ZC3H14zinc finger CCCH-type containing 14 (204216_s_at), score: -0.4 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: -0.72 ZFRzinc finger RNA binding protein (33148_at), score: -0.38 ZMYND11zinc finger, MYND domain containing 11 (202137_s_at), score: -0.78 ZNF137zinc finger protein 137 (207394_at), score: 0.83 ZNF195zinc finger protein 195 (204234_s_at), score: -0.38 ZNF26zinc finger protein 26 (219595_at), score: 0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |