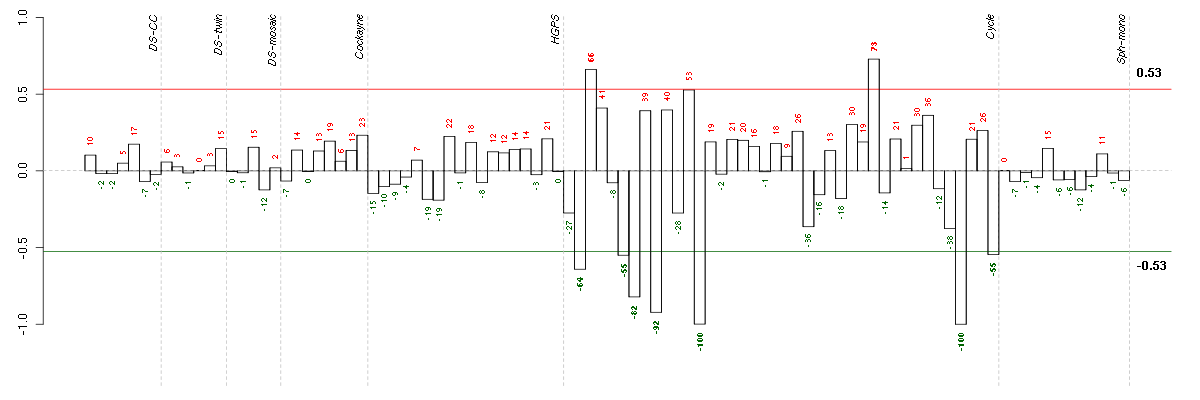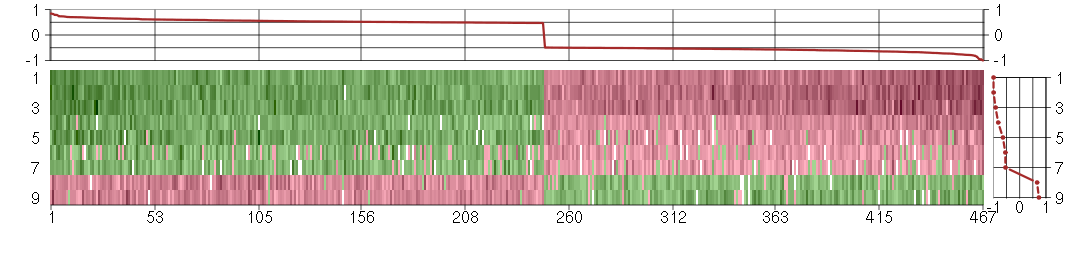

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.55 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.6 ABHD5abhydrolase domain containing 5 (218739_at), score: -0.51 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.56 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.54 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.67 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.61 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.8 ADAadenosine deaminase (204639_at), score: -0.54 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.61 ADARB1adenosine deaminase, RNA-specific, B1 (RED1 homolog rat) (203865_s_at), score: -0.49 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.51 ADORA2Badenosine A2b receptor (205891_at), score: 0.7 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.5 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.51 AHI1Abelson helper integration site 1 (221569_at), score: -0.68 AIM1absent in melanoma 1 (212543_at), score: 0.7 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: 0.51 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.57 ANXA10annexin A10 (210143_at), score: -0.69 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: -0.51 APOL6apolipoprotein L, 6 (219716_at), score: 0.5 APPamyloid beta (A4) precursor protein (200602_at), score: -0.6 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.57 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (211935_at), score: 0.55 ARSJarylsulfatase family, member J (219973_at), score: -0.57 ATF5activating transcription factor 5 (204999_s_at), score: 0.48 ATMataxia telangiectasia mutated (210858_x_at), score: -0.51 ATN1atrophin 1 (40489_at), score: 0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.67 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.52 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: -0.5 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.53 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.57 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.55 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.61 BCL2L1BCL2-like 1 (215037_s_at), score: 0.55 BMP6bone morphogenetic protein 6 (206176_at), score: -0.66 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.6 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.63 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.62 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.62 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.49 C14orf104chromosome 14 open reading frame 104 (219166_at), score: -0.5 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.7 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.59 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.55 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.51 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.55 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.48 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.69 C20orf20chromosome 20 open reading frame 20 (218586_at), score: 0.51 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: 0.48 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.55 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.73 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.6 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.51 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: -0.51 CABYRcalcium binding tyrosine-(Y)-phosphorylation regulated (219928_s_at), score: 0.55 CALB2calbindin 2 (205428_s_at), score: -0.79 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.61 CAPN5calpain 5 (205166_at), score: 0.53 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: 0.47 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.66 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.53 CCNFcyclin F (204826_at), score: 0.53 CD248CD248 molecule, endosialin (219025_at), score: 0.49 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.51 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.63 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: 0.52 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.66 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.6 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.49 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.68 CDCA3cell division cycle associated 3 (221436_s_at), score: 0.54 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.66 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.68 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: 0.5 CDKN3cyclin-dependent kinase inhibitor 3 (209714_s_at), score: 0.53 CENPEcentromere protein E, 312kDa (205046_at), score: 0.48 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: 0.47 CHEK2CHK2 checkpoint homolog (S. pombe) (210416_s_at), score: 0.48 CHKAcholine kinase alpha (204233_s_at), score: 0.51 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.53 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.62 CICcapicua homolog (Drosophila) (212784_at), score: 0.63 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.65 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.61 CLN5ceroid-lipofuscinosis, neuronal 5 (204084_s_at), score: -0.6 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.58 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.59 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.66 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.49 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.84 CScitrate synthase (208660_at), score: 0.61 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.56 CUX1cut-like homeobox 1 (214743_at), score: -0.57 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.55 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.53 DAXXdeath-domain associated protein (201763_s_at), score: 0.5 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.51 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.49 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.55 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: 0.53 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.6 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.51 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: -0.54 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.71 DERL2Der1-like domain family, member 2 (218333_at), score: -0.56 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.52 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.67 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.64 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.63 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.85 DNAJC1DnaJ (Hsp40) homolog, subfamily C, member 1 (218409_s_at), score: -0.49 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.5 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.7 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.52 DOCK9dedicator of cytokinesis 9 (212538_at), score: -0.52 DOPEY1dopey family member 1 (40612_at), score: -0.56 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: 0.52 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.49 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.51 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.62 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.54 EAF2ELL associated factor 2 (219551_at), score: -0.67 EBPemopamil binding protein (sterol isomerase) (213789_at), score: 0.49 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.69 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.67 EI24etoposide induced 2.4 mRNA (208289_s_at), score: -0.58 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.58 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.65 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.5 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.5 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.52 EPHA4EPH receptor A4 (206114_at), score: -0.63 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.5 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.65 EVI1ecotropic viral integration site 1 (221884_at), score: -0.53 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.51 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.55 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.67 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.53 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: 0.5 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.59 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: -0.57 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.49 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.79 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.51 FICDFIC domain containing (219910_at), score: -0.65 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.63 FLOT1flotillin 1 (210142_x_at), score: 0.5 FNBP1formin binding protein 1 (212288_at), score: 0.5 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.54 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.51 FOSL1FOS-like antigen 1 (204420_at), score: 0.57 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.59 FOXF1forkhead box F1 (205935_at), score: 0.49 FOXK2forkhead box K2 (203064_s_at), score: 0.7 FOXM1forkhead box M1 (202580_x_at), score: 0.58 FRG1FSHD region gene 1 (204145_at), score: -0.57 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.57 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.59 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.54 GALK2galactokinase 2 (205219_s_at), score: -0.5 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.77 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.51 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: -0.54 GHRgrowth hormone receptor (205498_at), score: -0.71 GKglycerol kinase (207387_s_at), score: -0.65 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.82 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.73 GMPRguanosine monophosphate reductase (204187_at), score: 0.61 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: -0.54 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.61 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.57 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: 0.6 GSK3Bglycogen synthase kinase 3 beta (209945_s_at), score: 0.49 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.6 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.5 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: 0.48 H1F0H1 histone family, member 0 (208886_at), score: 0.59 H1FXH1 histone family, member X (204805_s_at), score: 0.69 HAB1B1 for mucin (215778_x_at), score: -0.51 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.65 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.5 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.6 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.51 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.53 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.51 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.59 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.56 HRH1histamine receptor H1 (205580_s_at), score: 0.49 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.78 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.7 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.56 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: -0.5 ICAM3intercellular adhesion molecule 3 (204949_at), score: -0.5 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.54 IL33interleukin 33 (209821_at), score: -0.58 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.55 INSIG2insulin induced gene 2 (209566_at), score: -0.55 IRF1interferon regulatory factor 1 (202531_at), score: 0.54 IRS1insulin receptor substrate 1 (204686_at), score: -0.58 ISOC1isochorismatase domain containing 1 (218170_at), score: -0.52 ITGA1integrin, alpha 1 (214660_at), score: -0.64 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.61 JUNDjun D proto-oncogene (203751_x_at), score: 0.59 JUPjunction plakoglobin (201015_s_at), score: 0.62 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.96 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.57 KIAA0562KIAA0562 (204075_s_at), score: -0.6 KIAA1305KIAA1305 (220911_s_at), score: 0.57 KIAA1462KIAA1462 (213316_at), score: 0.57 KIF20Akinesin family member 20A (218755_at), score: 0.48 KLHL21kelch-like 21 (Drosophila) (203068_at), score: -0.55 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.73 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.66 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.61 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.59 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.49 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.53 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.64 LMNB2lamin B2 (216952_s_at), score: 0.53 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.64 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.65 LOC391132similar to hCG2041276 (216177_at), score: -0.59 LOC399491LOC399491 protein (214035_x_at), score: 0.58 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.55 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.57 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.56 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.72 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.69 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.51 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.5 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.59 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.56 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.52 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.55 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.48 MC4Rmelanocortin 4 receptor (221467_at), score: -0.65 MFAP3microfibrillar-associated protein 3 (213123_at), score: -0.53 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.76 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.52 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.6 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.55 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.53 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.56 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.51 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.58 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.52 NCALDneurocalcin delta (211685_s_at), score: -0.62 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.57 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.54 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.67 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.55 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.5 NEFLneurofilament, light polypeptide (221805_at), score: -0.61 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NEK2NIMA (never in mitosis gene a)-related kinase 2 (204641_at), score: 0.5 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.6 NF1neurofibromin 1 (211094_s_at), score: 0.64 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.6 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.63 NMIN-myc (and STAT) interactor (203964_at), score: 0.5 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.51 NOX4NADPH oxidase 4 (219773_at), score: -0.67 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.68 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.48 NPTX1neuronal pentraxin I (204684_at), score: -0.63 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.54 NRXN3neurexin 3 (205795_at), score: -0.64 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.51 NUP160nucleoporin 160kDa (212709_at), score: -0.55 NXPH3neurexophilin 3 (221991_at), score: -0.53 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.49 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.59 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.5 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.82 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.6 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.78 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.7 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.51 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.68 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.56 PARVBparvin, beta (37965_at), score: 0.56 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: 0.5 PCDH9protocadherin 9 (219737_s_at), score: -0.94 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.49 PCNXpecanex homolog (Drosophila) (213159_at), score: 0.49 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.58 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.56 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.65 PDE7Bphosphodiesterase 7B (220343_at), score: -0.56 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.49 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.48 PHLDA1pleckstrin homology-like domain, family A, member 1 (217997_at), score: 0.49 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (209625_at), score: -0.5 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.54 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.54 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.51 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.6 PKN1protein kinase N1 (202161_at), score: 0.53 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.51 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.59 PLCD1phospholipase C, delta 1 (205125_at), score: 0.53 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.64 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.55 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.56 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.49 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.52 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.49 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.52 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.54 PRKCDprotein kinase C, delta (202545_at), score: 0.53 PRKCHprotein kinase C, eta (218764_at), score: 0.51 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.76 PSMA2proteasome (prosome, macropain) subunit, alpha type, 2 (201316_at), score: -0.5 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.52 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.53 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.57 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.56 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.54 PXNpaxillin (211823_s_at), score: 0.57 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.74 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.51 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.53 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.54 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: -0.54 RANBP6RAN binding protein 6 (213019_at), score: -0.52 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.51 RBM15BRNA binding motif protein 15B (202689_at), score: 0.56 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.79 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.57 RGS17regulator of G-protein signaling 17 (220334_at), score: 0.52 RNF11ring finger protein 11 (208924_at), score: -0.6 RNF121ring finger protein 121 (219021_at), score: -0.51 RNF19Bring finger protein 19B (213038_at), score: 0.52 RNF220ring finger protein 220 (219988_s_at), score: 0.66 RNF44ring finger protein 44 (203286_at), score: 0.53 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.57 RP4-691N24.1ninein-like (207705_s_at), score: 0.57 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.58 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.78 RPL39Lribosomal protein L39-like (210115_at), score: 0.49 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.56 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.65 RPS25ribosomal protein S25 (200091_s_at), score: -0.53 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.48 RSRC1arginine/serine-rich coiled-coil 1 (219507_at), score: 0.47 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.5 SAP30LSAP30-like (219129_s_at), score: 0.72 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.59 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.59 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.71 SCARA3scavenger receptor class A, member 3 (219416_at), score: 0.53 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.63 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.48 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.5 SEPHS1selenophosphate synthetase 1 (208939_at), score: 0.64 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.7 SF1splicing factor 1 (208313_s_at), score: 0.73 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.51 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.47 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.5 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.55 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.51 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.52 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.6 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.47 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.51 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: 0.49 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.48 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.73 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.51 SLC39A7solute carrier family 39 (zinc transporter), member 7 (202667_s_at), score: -0.56 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.61 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.6 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.72 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.62 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.52 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.65 SMAD3SMAD family member 3 (218284_at), score: 0.67 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: 0.47 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.54 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.48 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.52 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.47 SNIP1Smad nuclear interacting protein 1 (219409_at), score: 0.53 SNNstannin (218032_at), score: 0.67 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.57 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.48 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.48 SSTR1somatostatin receptor 1 (208482_at), score: -0.6 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.5 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.47 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.49 STIM1stromal interaction molecule 1 (202764_at), score: 0.61 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.5 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.75 SYNE2spectrin repeat containing, nuclear envelope 2 (202761_s_at), score: 0.5 SYNJ1synaptojanin 1 (212990_at), score: -0.64 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.57 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.53 TARDBPTAR DNA binding protein (200020_at), score: 0.55 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (219771_at), score: -0.58 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.54 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.55 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.63 THADAthyroid adenoma associated (54632_at), score: -0.53 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.65 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.49 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.72 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.63 TLR3toll-like receptor 3 (206271_at), score: 0.54 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.58 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.53 TMEM104transmembrane protein 104 (220097_s_at), score: -0.65 TMEM2transmembrane protein 2 (218113_at), score: -0.59 TMEM30Btransmembrane protein 30B (213285_at), score: 0.47 TMSB15Bthymosin beta 15B (214051_at), score: 0.65 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.61 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.52 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.55 TP53tumor protein p53 (201746_at), score: 0.7 TPBGtrophoblast glycoprotein (203476_at), score: 0.54 TRAF5TNF receptor-associated factor 5 (204352_at), score: -0.54 TRAPPC2trafficking protein particle complex 2 (219351_at), score: -0.55 TRIM21tripartite motif-containing 21 (204804_at), score: 0.68 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.62 TROAPtrophinin associated protein (tastin) (204649_at), score: 0.52 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.66 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.53 TSKUtsukushin (218245_at), score: 0.6 TSPAN31tetraspanin 31 (203227_s_at), score: -0.49 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.54 TXLNAtaxilin alpha (212300_at), score: 0.5 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.5 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.48 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.59 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.51 ULBP2UL16 binding protein 2 (221291_at), score: -0.57 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.61 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.57 VGLL3vestigial like 3 (Drosophila) (220327_at), score: -0.5 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.53 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.5 WDR4WD repeat domain 4 (221632_s_at), score: -0.55 WDR42AWD repeat domain 42A (202249_s_at), score: 0.55 WDR6WD repeat domain 6 (217734_s_at), score: 0.73 WDR62WD repeat domain 62 (215218_s_at), score: 0.52 XAB2XPA binding protein 2 (218110_at), score: 0.48 XBP1X-box binding protein 1 (200670_at), score: -0.5 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.73 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.52 YTHDC2YTH domain containing 2 (213077_at), score: -0.59 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.48 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.56 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.7 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: -0.56 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.51 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.5 ZNF20zinc finger protein 20 (213916_at), score: 0.48 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.62 ZNF395zinc finger protein 395 (218149_s_at), score: 0.47 ZNF706zinc finger protein 706 (218059_at), score: 0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |