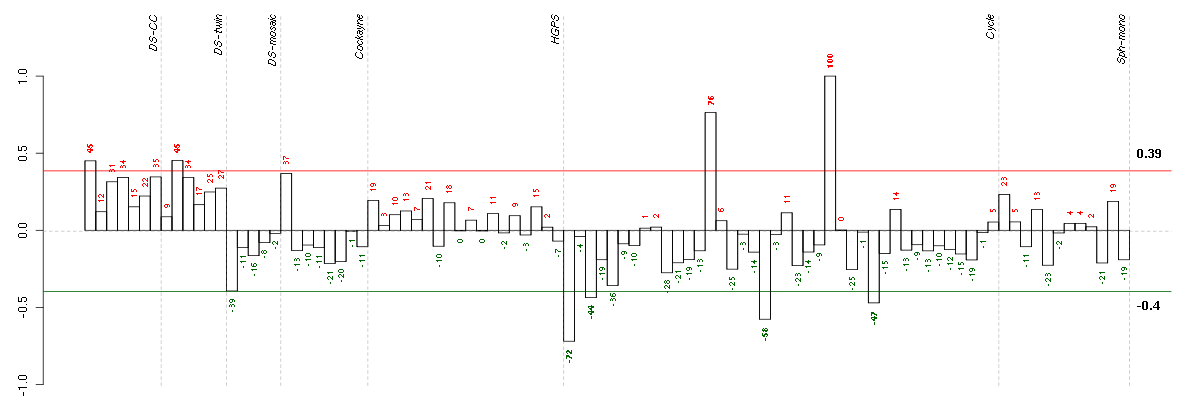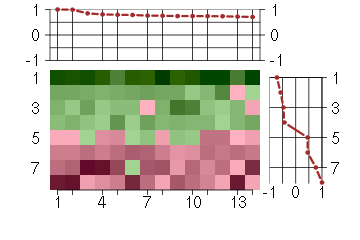

Under-expression is coded with green,
over-expression with red color.



ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: 0.71 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: 0.79 GPR12G protein-coupled receptor 12 (214558_at), score: 0.74 HIST1H2ALhistone cluster 1, H2al (214554_at), score: 0.99 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.74 LHX3LIM homeobox 3 (221670_s_at), score: 0.73 MAGI1membrane associated guanylate kinase, WW and PDZ domain containing 1 (206144_at), score: 0.72 MED31mediator complex subunit 31 (219318_x_at), score: 0.74 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: 1 MYO5Cmyosin VC (218966_at), score: 0.85 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.76 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: 0.76 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.79 TAAR2trace amine associated receptor 2 (221394_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |