
Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
protein serine/threonine kinase activity
Catalysis of the reaction: ATP + a protein serine/threonine = ADP + protein serine/threonine phosphate.
protein kinase C activity
Catalysis of the reaction: ATP + a protein = ADP + a phosphoprotein, with a requirement for diacylglycerol.
kinase activity
Catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring phosphorus-containing groups
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to another (acceptor).
phosphotransferase activity, alcohol group as acceptor
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to an alcohol group (acceptor).
all
This term is the most general term possible
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
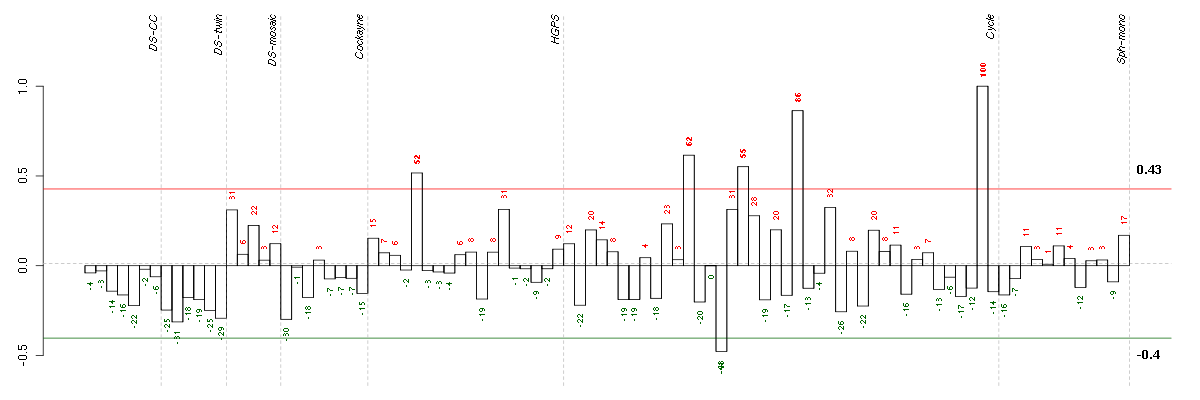
BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.68 HOOK1hook homolog 1 (Drosophila) (219976_at), score: -0.88 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: 1 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: -0.82 PRKCBprotein kinase C, beta (209685_s_at), score: -0.78 PRKCDprotein kinase C, delta (202545_at), score: 0.71 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.69 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.69 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.7 XYLBxylulokinase homolog (H. influenzae) (214776_x_at), score: -0.76 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |