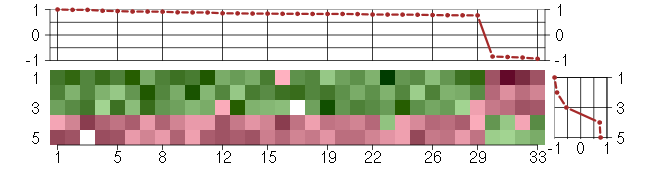

Under-expression is coded with green,
over-expression with red color.



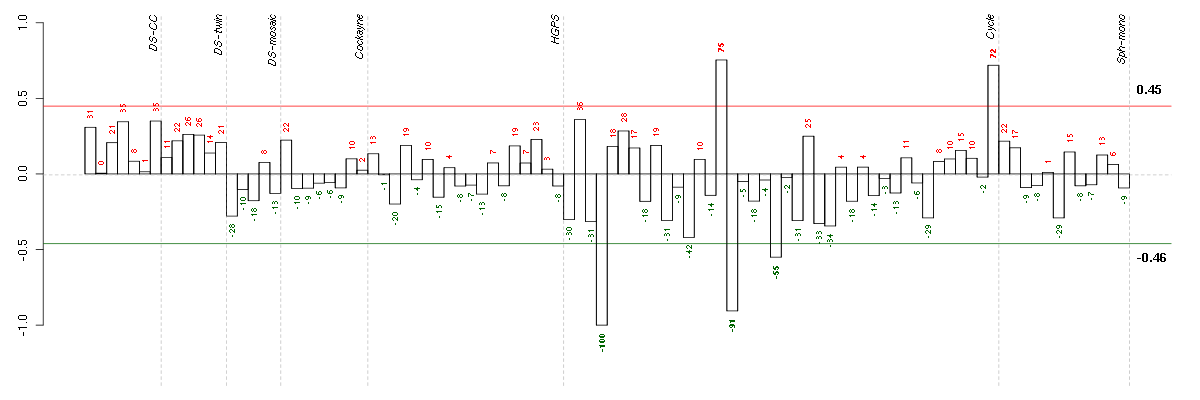
ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.83 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.84 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.83 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.85 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.94 CYLC2cylicin, basic protein of sperm head cytoskeleton 2 (207780_at), score: 0.83 DSPPdentin sialophosphoprotein (221681_s_at), score: 1 DUSP9dual specificity phosphatase 9 (205777_at), score: 0.91 F11coagulation factor XI (206610_s_at), score: 0.92 FLCNfolliculin (215645_at), score: 0.8 GANgigaxonin (220124_at), score: -0.93 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.99 GPR144G protein-coupled receptor 144 (216289_at), score: 0.88 GPR32G protein-coupled receptor 32 (221469_at), score: 0.8 HAB1B1 for mucin (215778_x_at), score: 0.89 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.77 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.82 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.99 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.79 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.85 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.77 PCDH24protocadherin 24 (220186_s_at), score: 0.83 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.84 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.78 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.88 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.8 RBM38RNA binding motif protein 38 (212430_at), score: 0.83 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.78 SLCO2A1solute carrier organic anion transporter family, member 2A1 (204368_at), score: 0.84 SNTG1syntrophin, gamma 1 (220405_at), score: -0.88 SPATA1spermatogenesis associated 1 (221057_at), score: 0.92 TULP2tubby like protein 2 (206733_at), score: 0.95 ZNF460zinc finger protein 460 (216279_at), score: -0.86
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |