
Under-expression is coded with green,
over-expression with red color.



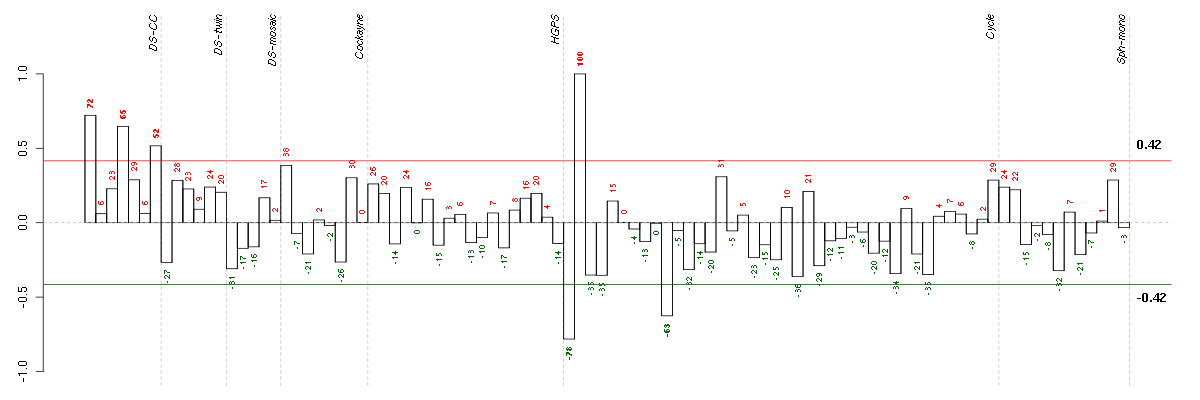
ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.87 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.87 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.98 LOC80054hypothetical LOC80054 (220465_at), score: 0.88 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.99 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.95 PGK2phosphoglycerate kinase 2 (217009_at), score: 0.9 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 1 TRA@T cell receptor alpha locus (216540_at), score: 0.91 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.96
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |