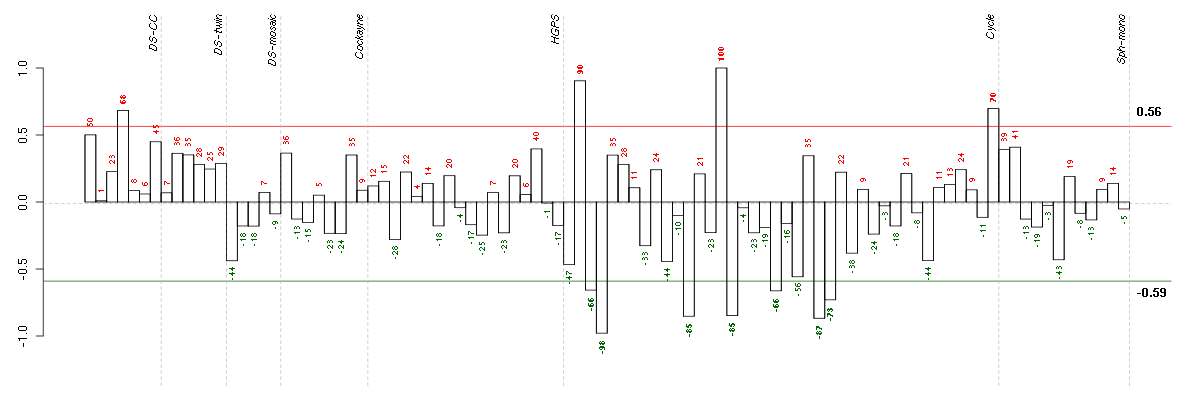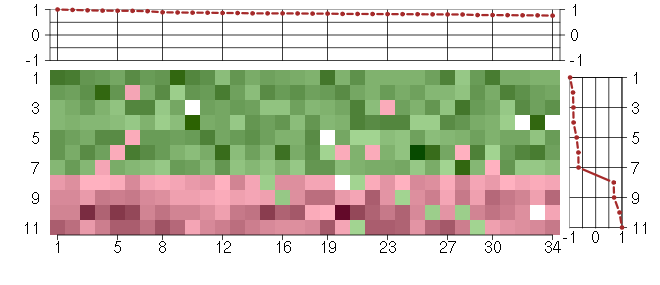

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.83 ADORA1adenosine A1 receptor (216220_s_at), score: 0.86 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.82 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.95 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.8 CYTH4cytohesin 4 (219183_s_at), score: 0.86 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.83 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.81 F11coagulation factor XI (206610_s_at), score: 0.77 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.95 FLCNfolliculin (215645_at), score: 0.84 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.76 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.77 GPR144G protein-coupled receptor 144 (216289_at), score: 0.88 HAB1B1 for mucin (215778_x_at), score: 0.98 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.97 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.93 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 1 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: 0.89 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.81 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.87 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.81 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.78 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.85 OPRL1opiate receptor-like 1 (206564_at), score: 0.85 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.82 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.96 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.88 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.78 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.78 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.82 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.85 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.85 TULP2tubby like protein 2 (206733_at), score: 0.84
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |