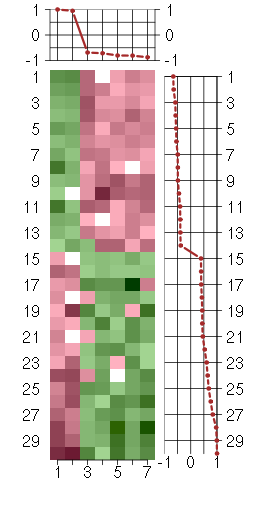

Under-expression is coded with green,
over-expression with red color.



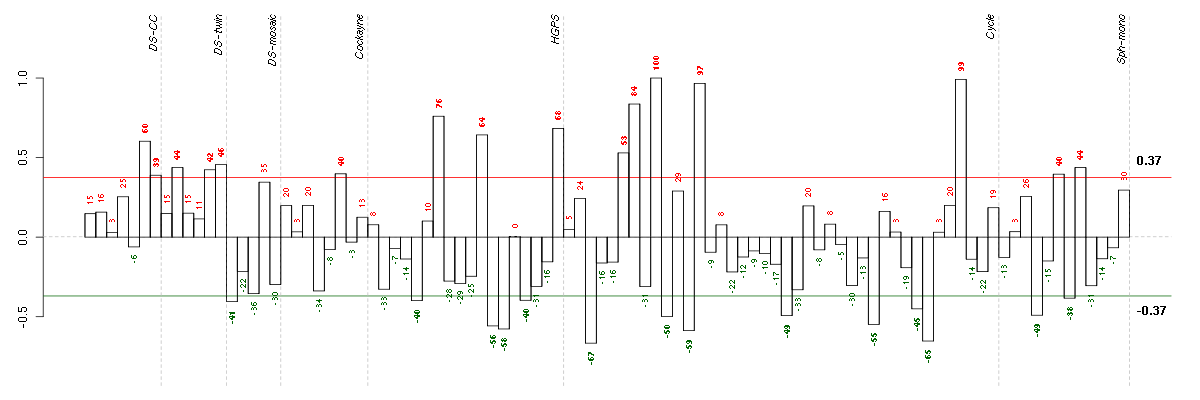
CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.8 ECM1extracellular matrix protein 1 (209365_s_at), score: -0.81 KAL1Kallmann syndrome 1 sequence (205206_at), score: 1 NOX4NADPH oxidase 4 (219773_at), score: 0.95 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: -0.72 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.69 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.87
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |