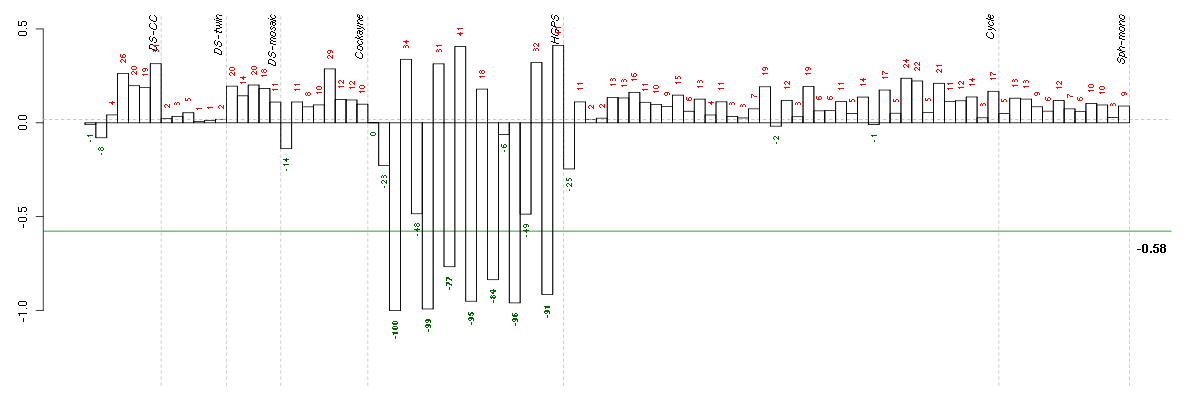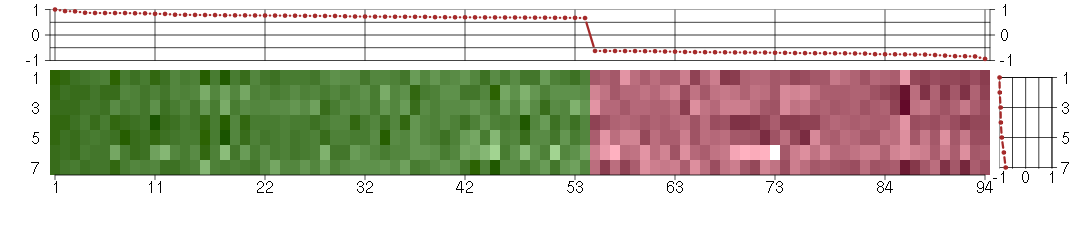

Under-expression is coded with green,
over-expression with red color.

skeletal system development
The process whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure. The skeleton is the bony framework of the body in vertebrates (endoskeleton) or the hard outer envelope of insects (exoskeleton or dermoskeleton).
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
bone mineralization
The deposition of calcium phosphate in bone tissue.
biomineral formation
Formation of hard tissues that consist mainly of inorganic compounds, and also contain a small amounts of organic matrices that are believed to play important roles in their formation.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.
bone mineralization
The deposition of calcium phosphate in bone tissue.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.93 AUTS2autism susceptibility candidate 2 (212599_at), score: -0.72 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.79 C1orf54chromosome 1 open reading frame 54 (219506_at), score: 0.69 C1QTNF3C1q and tumor necrosis factor related protein 3 (220988_s_at), score: -0.76 CA12carbonic anhydrase XII (203963_at), score: -0.7 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: -0.94 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: 0.69 CCR10chemokine (C-C motif) receptor 10 (220565_at), score: 0.68 CDH4cadherin 4, type 1, R-cadherin (retinal) (220227_at), score: -0.63 CLGNcalmegin (205830_at), score: -0.68 COL4A1collagen, type IV, alpha 1 (211981_at), score: 0.67 CTSCcathepsin C (201487_at), score: 0.71 CYP1B1cytochrome P450, family 1, subfamily B, polypeptide 1 (202436_s_at), score: 0.78 DMDdystrophin (203881_s_at), score: 0.71 DNM1dynamin 1 (215116_s_at), score: -0.63 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: 0.77 EFEMP1EGF-containing fibulin-like extracellular matrix protein 1 (201842_s_at), score: 0.67 EIF2S3eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa (205321_at), score: 0.68 EMX2empty spiracles homeobox 2 (221950_at), score: -0.65 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: -0.75 EPHA5EPH receptor A5 (215664_s_at), score: 0.86 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: 0.86 FHL1four and a half LIM domains 1 (201539_s_at), score: 0.69 FHOD3formin homology 2 domain containing 3 (218980_at), score: -0.75 FSTfollistatin (204948_s_at), score: 0.72 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.71 GPERG protein-coupled estrogen receptor 1 (210640_s_at), score: -0.68 GPR116G protein-coupled receptor 116 (212950_at), score: 0.78 HMOX1heme oxygenase (decycling) 1 (203665_at), score: -0.72 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.79 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.77 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.72 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: 0.85 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: -0.77 KRT18keratin 18 (201596_x_at), score: 0.83 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.7 LIPGlipase, endothelial (219181_at), score: 0.79 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: 0.76 LPHN2latrophilin 2 (206953_s_at), score: 0.86 LPXNleupaxin (216250_s_at), score: -0.71 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.84 LRRC17leucine rich repeat containing 17 (205381_at), score: 0.75 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.69 MBPmyelin basic protein (210136_at), score: 0.74 MEOX2mesenchyme homeobox 2 (206201_s_at), score: 1 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.87 MGPmatrix Gla protein (202291_s_at), score: 0.85 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: -0.63 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: -0.63 MSX1msh homeobox 1 (205932_s_at), score: -0.67 MT1Mmetallothionein 1M (217546_at), score: -0.73 MYCT1myc target 1 (220471_s_at), score: 0.67 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.7 NDUFA4L2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (218484_at), score: 0.94 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.76 NID2nidogen 2 (osteonidogen) (204114_at), score: 0.72 NKX3-1NK3 homeobox 1 (209706_at), score: -0.83 NOVnephroblastoma overexpressed gene (214321_at), score: 0.7 NRG1neuregulin 1 (206343_s_at), score: -0.64 NRIP3nuclear receptor interacting protein 3 (219557_s_at), score: -0.63 OLFM1olfactomedin 1 (213131_at), score: -0.69 OLFML3olfactomedin-like 3 (218162_at), score: -0.65 PDE10Aphosphodiesterase 10A (205501_at), score: 0.75 PLA2G16phospholipase A2, group XVI (209581_at), score: 0.79 PLXND1plexin D1 (38671_at), score: 0.75 PNMA2paraneoplastic antigen MA2 (209598_at), score: 0.76 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.77 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (218292_s_at), score: 0.7 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.71 PTX3pentraxin-related gene, rapidly induced by IL-1 beta (206157_at), score: 0.73 RAB15RAB15, member RAS onocogene family (221810_at), score: -0.69 RELNreelin (205923_at), score: 0.77 RGS7regulator of G-protein signaling 7 (206290_s_at), score: 0.68 RORARAR-related orphan receptor A (210426_x_at), score: -0.69 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.69 SETBP1SET binding protein 1 (205933_at), score: -0.71 SFRP1secreted frizzled-related protein 1 (202036_s_at), score: -0.63 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.68 SLC25A4solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 (202825_at), score: 0.71 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: -0.81 SLCO3A1solute carrier organic anion transporter family, member 3A1 (219229_at), score: -0.71 SRGNserglycin (201859_at), score: 0.83 SSX2IPsynovial sarcoma, X breakpoint 2 interacting protein (203016_s_at), score: -0.66 STMN2stathmin-like 2 (203000_at), score: -0.67 SUSD5sushi domain containing 5 (214954_at), score: 0.7 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: 0.77 TLR4toll-like receptor 4 (221060_s_at), score: 0.67 TMEM158transmembrane protein 158 (213338_at), score: -0.83 TNXAtenascin XA pseudogene (213451_x_at), score: 0.76 TNXBtenascin XB (216333_x_at), score: 0.75 TUFT1tuftelin 1 (205807_s_at), score: 0.72 ZNF423zinc finger protein 423 (214761_at), score: 0.86 ZNF536zinc finger protein 536 (206403_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |