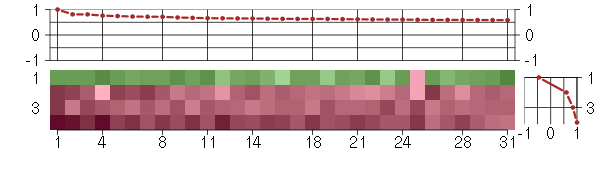

Under-expression is coded with green,
over-expression with red color.



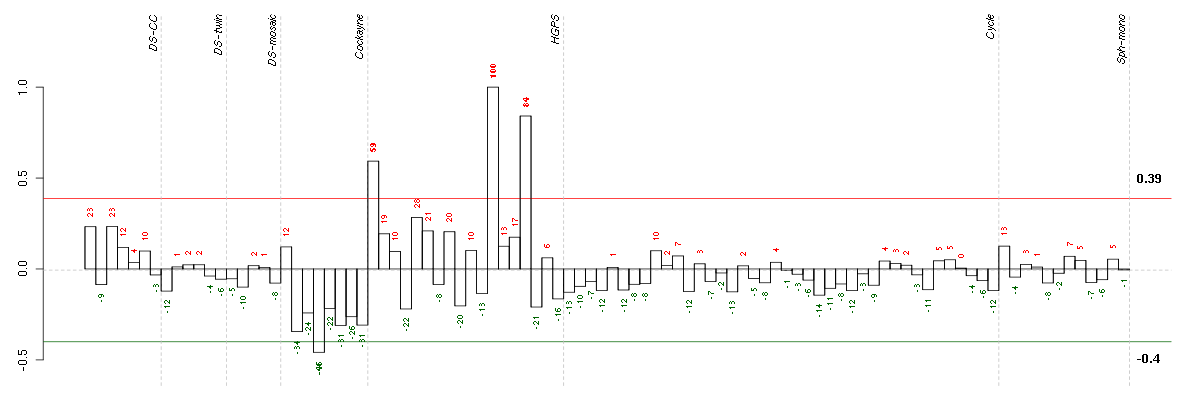
ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: 0.64 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.58 BIN3bridging integrator 3 (222199_s_at), score: 0.59 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.81 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 1 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.59 CKBcreatine kinase, brain (200884_at), score: 0.63 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.6 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.63 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.74 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.65 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.62 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.59 HOXA11homeobox A11 (213823_at), score: 0.62 HOXA9homeobox A9 (214651_s_at), score: 0.66 IMAASLC7A5 pseudogene (208118_x_at), score: 0.67 INHBBinhibin, beta B (205258_at), score: 0.71 IRX5iroquois homeobox 5 (210239_at), score: 0.65 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: 0.63 KRT14keratin 14 (209351_at), score: 0.59 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.61 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.76 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.6 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.61 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.72 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.72 RUNX3runt-related transcription factor 3 (204198_s_at), score: 0.69 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.58 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.63 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.81 TBX5T-box 5 (207155_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |