
Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

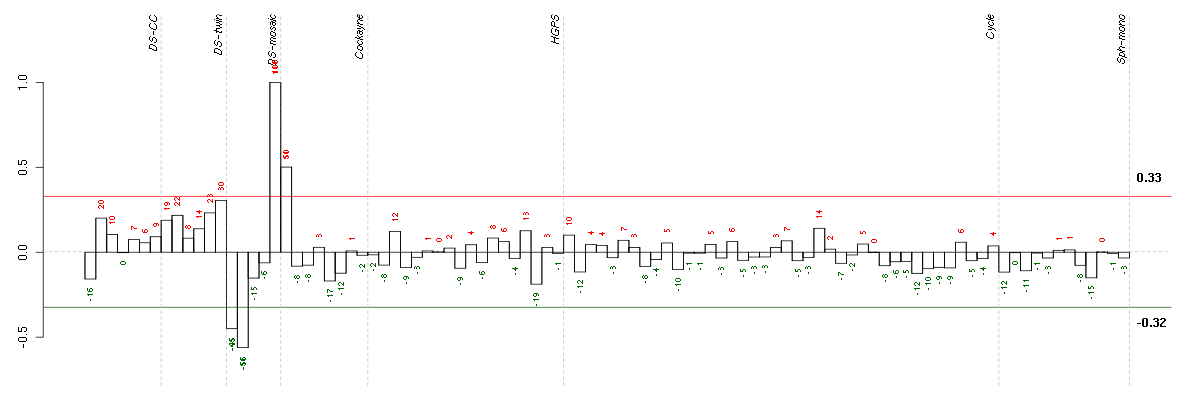
ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.76 APOL1apolipoprotein L, 1 (209546_s_at), score: -0.72 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.73 CA11carbonic anhydrase XI (209726_at), score: -0.84 CFIcomplement factor I (203854_at), score: -0.75 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: -0.78 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: -0.84 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: -1 GPC4glypican 4 (204983_s_at), score: -0.79 HHEXhematopoietically expressed homeobox (215933_s_at), score: -0.85 HOXB9homeobox B9 (216417_x_at), score: 0.81 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.67 LAMA5laminin, alpha 5 (210150_s_at), score: -0.65 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: -0.67 NINJ1ninjurin 1 (203045_at), score: -0.68 PAPPA2pappalysin 2 (213332_at), score: -0.81 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: 0.91 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: -0.71 PKP2plakophilin 2 (207717_s_at), score: -0.84 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: -0.84 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.79 SCARB1scavenger receptor class B, member 1 (201819_at), score: -0.85 SCG2secretogranin II (chromogranin C) (204035_at), score: -0.89 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.73 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: -0.71 TMEM132Atransmembrane protein 132A (218834_s_at), score: -0.72 ZNF711zinc finger protein 711 (207781_s_at), score: -0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |