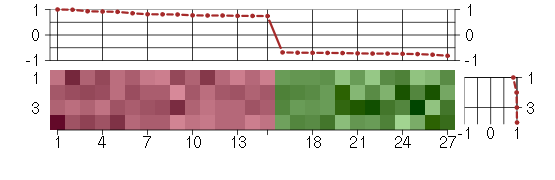

Under-expression is coded with green,
over-expression with red color.



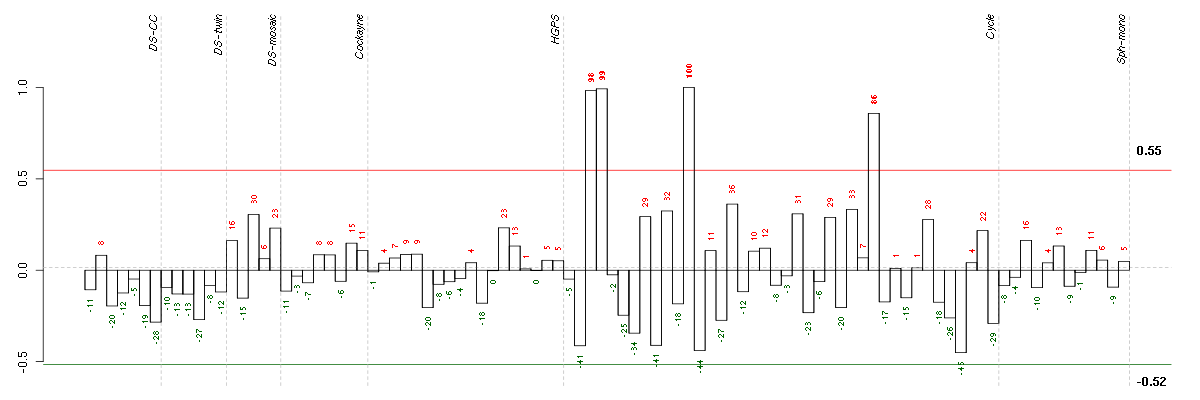
ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.69 CALB2calbindin 2 (205428_s_at), score: -0.7 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.77 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.75 EMCNendomucin (219436_s_at), score: 1 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.75 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.71 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.7 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.76 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.74 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.75 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.72 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: -0.74 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.81 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.8 NF1neurofibromin 1 (211094_s_at), score: 0.76 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.99 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.92 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.81 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.73 PXNpaxillin (211823_s_at), score: 0.77 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.91 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.93 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.81 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.85 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.68 ZNF652zinc finger protein 652 (205594_at), score: -0.73
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |