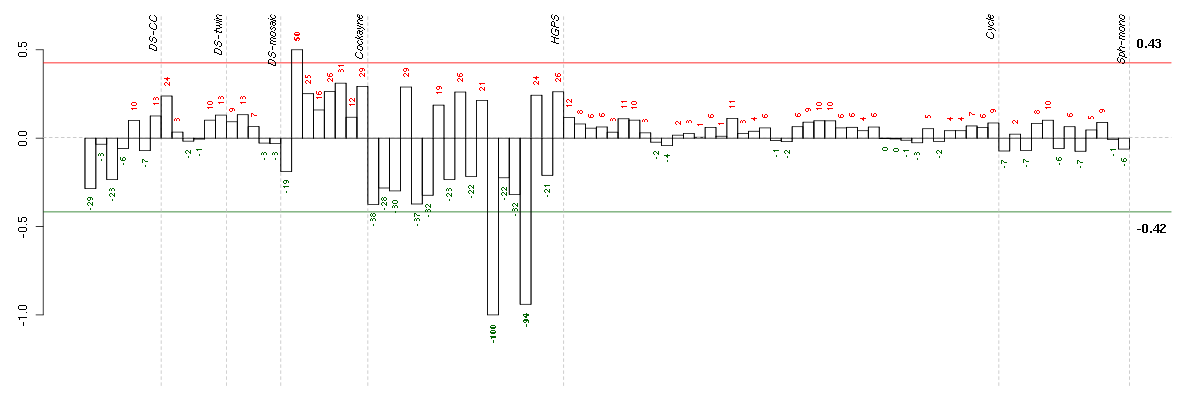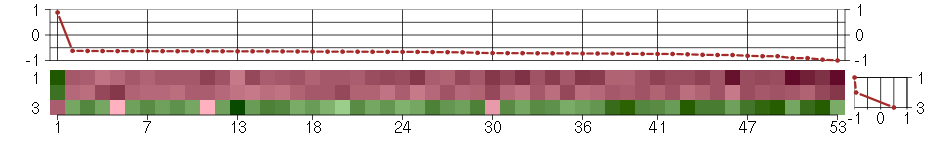

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: -0.72 ADCY7adenylate cyclase 7 (203741_s_at), score: -0.63 ANTXR1anthrax toxin receptor 1 (220092_s_at), score: -0.64 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.97 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: -1 CH25Hcholesterol 25-hydroxylase (206932_at), score: -0.7 CKBcreatine kinase, brain (200884_at), score: -0.78 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.71 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.65 COL8A2collagen, type VIII, alpha 2 (221900_at), score: -0.78 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.74 CPEcarboxypeptidase E (201117_s_at), score: -0.64 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: -0.63 EMX2empty spiracles homeobox 2 (221950_at), score: -0.83 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: -0.63 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: -0.73 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: -0.73 GRAMD4GRAM domain containing 4 (212856_at), score: -0.66 IER3immediate early response 3 (201631_s_at), score: 0.88 IL11RAinterleukin 11 receptor, alpha (204773_at), score: -0.67 IL1R1interleukin 1 receptor, type I (202948_at), score: -0.66 INHBBinhibin, beta B (205258_at), score: -0.73 IRX5iroquois homeobox 5 (210239_at), score: -0.91 ITPR1inositol 1,4,5-triphosphate receptor, type 1 (203710_at), score: -0.65 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.72 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: -0.64 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.71 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: -0.65 MEGF6multiple EGF-like-domains 6 (213942_at), score: -0.63 MSX1msh homeobox 1 (205932_s_at), score: -0.66 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.63 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.82 NCAM1neural cell adhesion molecule 1 (212843_at), score: -0.91 NTF3neurotrophin 3 (206706_at), score: -0.76 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: -0.74 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: -0.64 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: -0.68 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: -0.64 PSG6pregnancy specific beta-1-glycoprotein 6 (209738_x_at), score: -0.66 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: -0.64 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.72 RASIP1Ras interacting protein 1 (220027_s_at), score: -0.71 RUNX3runt-related transcription factor 3 (204198_s_at), score: -0.74 SIRPAsignal-regulatory protein alpha (202897_at), score: -0.63 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.66 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (220979_s_at), score: -0.64 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: -0.79 TBX5T-box 5 (207155_at), score: -0.68 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: -0.83 TLN2talin 2 (212703_at), score: -0.64 TSPAN13tetraspanin 13 (217979_at), score: -0.74 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: -0.62 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: -0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |