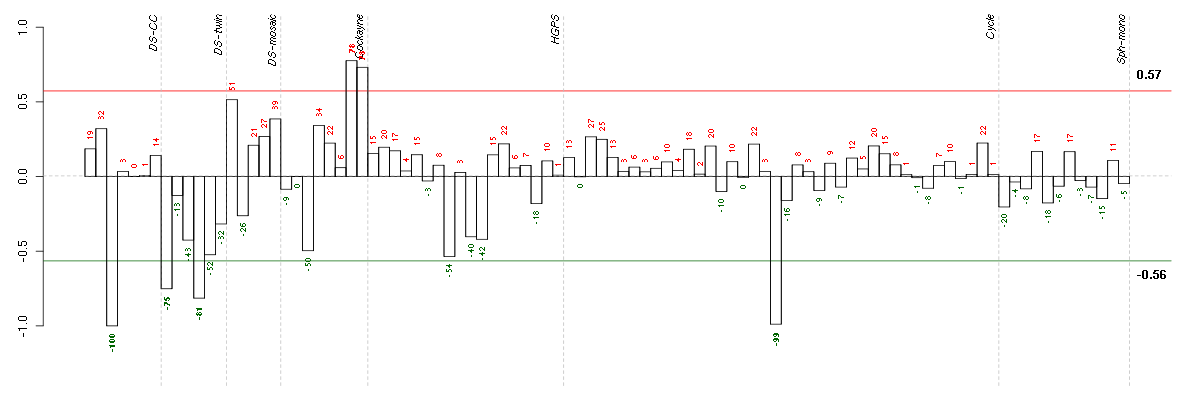

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-320/320abcd | 4.115e-02 | 5.714 | 18 | 372 |
ACLYATP citrate lyase (210337_s_at), score: 0.66 ADAM10ADAM metallopeptidase domain 10 (214895_s_at), score: 0.73 ARF1ADP-ribosylation factor 1 (208750_s_at), score: 0.67 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (216266_s_at), score: 0.67 ASPHaspartate beta-hydroxylase (205808_at), score: 0.66 ATP13A3ATPase type 13A3 (219558_at), score: 0.75 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.76 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.73 C20orf30chromosome 20 open reading frame 30 (220477_s_at), score: 0.65 CALUcalumenin (214845_s_at), score: 0.81 CANXcalnexin (208853_s_at), score: 0.72 CAPRIN1cell cycle associated protein 1 (200722_s_at), score: 0.69 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.66 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: 0.7 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.91 CLIC4chloride intracellular channel 4 (201559_s_at), score: 0.66 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.85 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.97 CPDcarboxypeptidase D (201942_s_at), score: 1 CYP51A1cytochrome P450, family 51, subfamily A, polypeptide 1 (216607_s_at), score: 0.79 DAZAP2DAZ associated protein 2 (212595_s_at), score: 0.8 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.99 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.99 DSTdystonin (212253_x_at), score: 0.66 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: 0.71 ERLIN1ER lipid raft associated 1 (202444_s_at), score: 0.71 EXOC5exocyst complex component 5 (218748_s_at), score: 0.7 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: 0.69 FKBP1AFK506 binding protein 1A, 12kDa (210186_s_at), score: 0.73 FN1fibronectin 1 (214701_s_at), score: 0.88 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: 0.63 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: 0.78 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.89 GPR137G protein-coupled receptor 137 (43934_at), score: 0.66 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.85 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: 0.65 HOXB6homeobox B6 (205366_s_at), score: 0.66 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.63 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: 0.7 KITLGKIT ligand (211124_s_at), score: 0.66 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: 0.77 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: 0.85 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.63 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: 0.85 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: 0.75 MAP3K7IP2mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (210284_s_at), score: 0.86 MAPK1mitogen-activated protein kinase 1 (208351_s_at), score: 0.74 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.71 MARCH7membrane-associated ring finger (C3HC4) 7 (202654_x_at), score: 0.69 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: 0.87 MAXMYC associated factor X (210734_x_at), score: 0.8 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: 0.78 NAB1NGFI-A binding protein 1 (EGR1 binding protein 1) (208047_s_at), score: 0.66 NID1nidogen 1 (202008_s_at), score: 0.69 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.78 OSMRoncostatin M receptor (205729_at), score: 0.7 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.72 PAPOLApoly(A) polymerase alpha (212720_at), score: 0.66 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.95 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.85 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.92 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: 0.78 PIP5K1Aphosphatidylinositol-4-phosphate 5-kinase, type I, alpha (211205_x_at), score: 0.65 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.67 PRPF4BPRP4 pre-mRNA processing factor 4 homolog B (yeast) (211090_s_at), score: 0.78 PRRX1paired related homeobox 1 (205991_s_at), score: 0.85 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.86 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.68 PXNpaxillin (211823_s_at), score: 0.75 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.95 RAB5CRAB5C, member RAS oncogene family (201156_s_at), score: 0.8 RANBP2RAN binding protein 2 (201711_x_at), score: 0.69 RANBP9RAN binding protein 9 (202583_s_at), score: 0.64 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: 0.67 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: 0.77 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: 0.69 SEPT11septin 11 (201308_s_at), score: 0.64 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.85 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.68 SPAG9sperm associated antigen 9 (206748_s_at), score: 0.66 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.67 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.71 SYPL1synaptophysin-like 1 (201259_s_at), score: 0.85 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.75 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.76 TJP1tight junction protein 1 (zona occludens 1) (214168_s_at), score: 0.64 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: 0.65 TWF1twinfilin, actin-binding protein, homolog 1 (Drosophila) (214007_s_at), score: 0.7 UBXN4UBX domain protein 4 (212008_at), score: 0.74 VAMP3vesicle-associated membrane protein 3 (cellubrevin) (201337_s_at), score: 0.75 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.98 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: 0.64 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: 0.68 ZMYND11zinc finger, MYND domain containing 11 (202137_s_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |