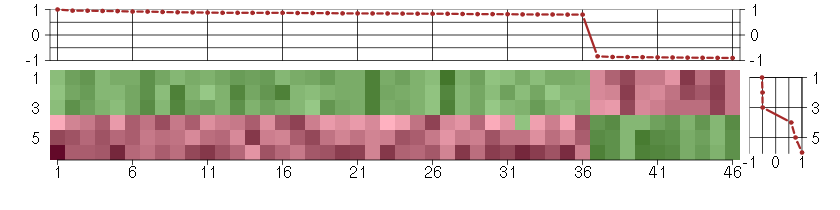

Under-expression is coded with green,
over-expression with red color.

response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

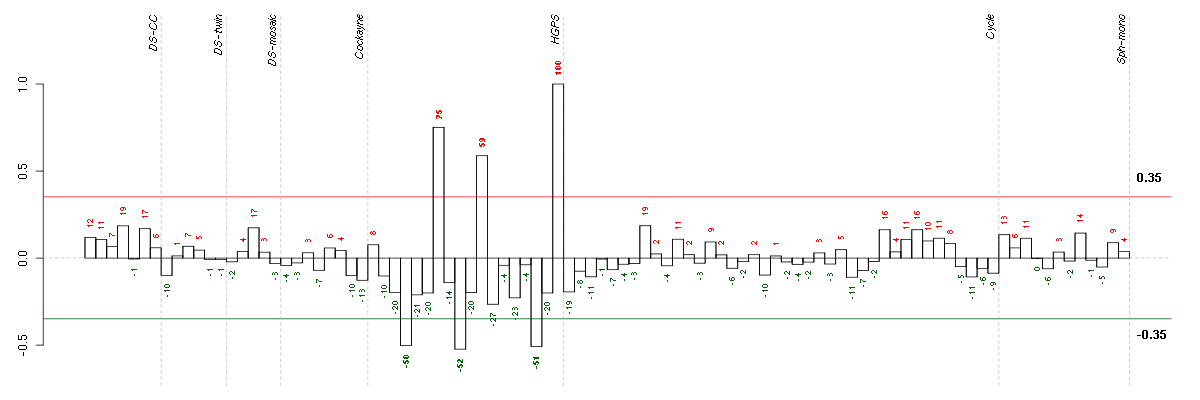
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.85 ADAMTSL3ADAMTS-like 3 (213974_at), score: 0.8 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.82 ANXA3annexin A3 (209369_at), score: 0.85 BNC2basonuclin 2 (220272_at), score: 0.87 C8orf84chromosome 8 open reading frame 84 (214725_at), score: 0.86 CD200CD200 molecule (209583_s_at), score: 0.94 CD34CD34 molecule (209543_s_at), score: 0.82 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.92 CFHcomplement factor H (213800_at), score: 0.88 CHRDL1chordin-like 1 (209763_at), score: 0.84 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.87 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.87 DSG2desmoglein 2 (217901_at), score: 0.87 EN1engrailed homeobox 1 (220559_at), score: -0.88 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (213541_s_at), score: 0.83 FBLN2fibulin 2 (203886_s_at), score: -0.86 FGL2fibrinogen-like 2 (204834_at), score: 1 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.81 GPR116G protein-coupled receptor 116 (212950_at), score: -0.9 GPR126G protein-coupled receptor 126 (213094_at), score: 0.89 GPR4G protein-coupled receptor 4 (206236_at), score: 0.86 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.84 HSPB8heat shock 22kDa protein 8 (221667_s_at), score: -0.88 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.84 IL18interleukin 18 (interferon-gamma-inducing factor) (206295_at), score: 0.8 IL1Ainterleukin 1, alpha (210118_s_at), score: 0.81 ITGA8integrin, alpha 8 (214265_at), score: -0.89 KCTD12potassium channel tetramerisation domain containing 12 (212192_at), score: 0.89 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: 0.87 LAMA4laminin, alpha 4 (202202_s_at), score: 0.83 LAMC2laminin, gamma 2 (202267_at), score: 0.86 LIPGlipase, endothelial (219181_at), score: 0.94 MYH10myosin, heavy chain 10, non-muscle (212372_at), score: 0.84 MYLKmyosin light chain kinase (202555_s_at), score: -0.84 NFIBnuclear factor I/B (209290_s_at), score: 0.82 NOX4NADPH oxidase 4 (219773_at), score: 0.92 PARP3poly (ADP-ribose) polymerase family, member 3 (209940_at), score: -0.9 PLXNA2plexin A2 (213030_s_at), score: 0.96 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: -0.87 RPP25ribonuclease P/MRP 25kDa subunit (219143_s_at), score: -0.89 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: 0.9 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: -0.87 TLR4toll-like receptor 4 (221060_s_at), score: 0.82 ZFHX4zinc finger homeobox 4 (219779_at), score: 0.95 ZNF323zinc finger protein 323 (222016_s_at), score: 0.8
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |