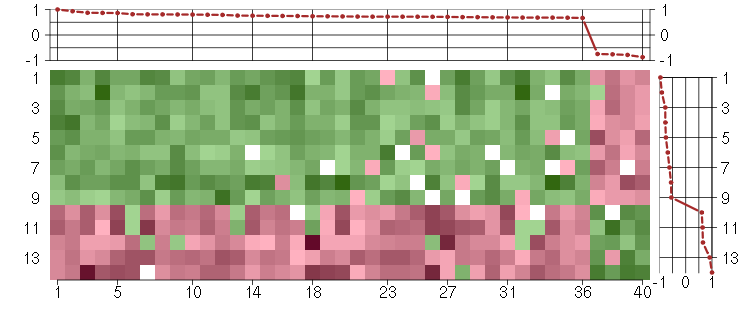

Under-expression is coded with green,
over-expression with red color.



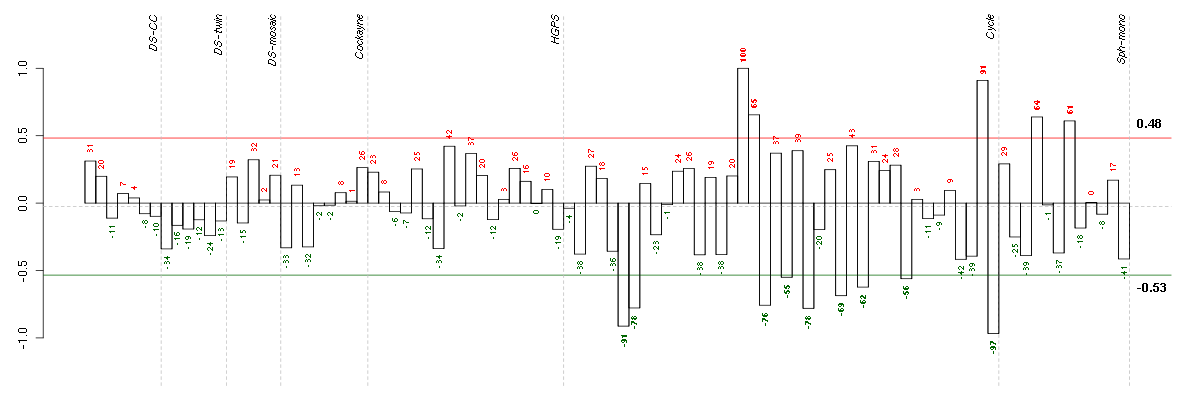
ANKRD28ankyrin repeat domain 28 (213035_at), score: -0.87 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.79 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.73 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.7 C20orf11chromosome 20 open reading frame 11 (218448_at), score: 0.68 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.73 CYTH1cytohesin 1 (202880_s_at), score: 0.86 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.74 DOK4docking protein 4 (209691_s_at), score: 0.75 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.8 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.93 FOXK2forkhead box K2 (203064_s_at), score: 1 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.72 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.71 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.68 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.87 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.81 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.68 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.81 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.72 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.73 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.69 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.81 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.76 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.72 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.79 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: -0.75 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.7 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.8 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.71 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.76 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.67 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.75 TSKUtsukushin (218245_at), score: 0.87 USP36ubiquitin specific peptidase 36 (220370_s_at), score: 0.72 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.68 WDR42AWD repeat domain 42A (202249_s_at), score: 0.72 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.76 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.69 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.78
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |