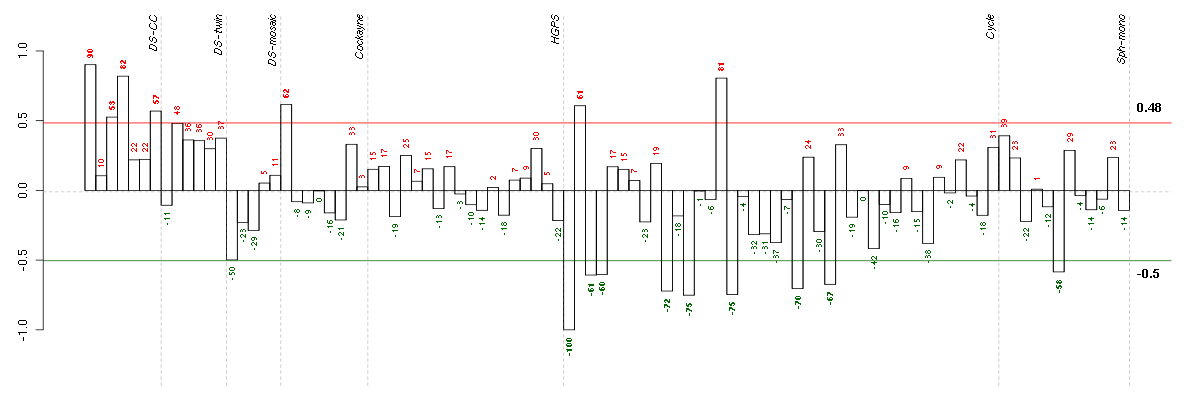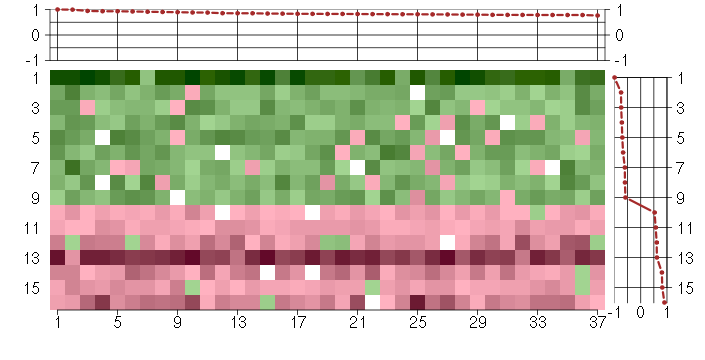

Under-expression is coded with green,
over-expression with red color.



ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.79 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.85 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: 0.82 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.83 CCDC9coiled-coil domain containing 9 (206257_at), score: 0.79 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.93 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.99 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.78 FGL1fibrinogen-like 1 (205305_at), score: 0.84 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.78 HAB1B1 for mucin (215778_x_at), score: 0.91 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.84 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.9 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.81 LOC80054hypothetical LOC80054 (220465_at), score: 0.85 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.94 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.83 MYO1Amyosin IA (211916_s_at), score: 0.88 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.82 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.76 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.8 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.79 PIPOXpipecolic acid oxidase (221605_s_at), score: 0.81 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: 0.93 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.92 RLN1relaxin 1 (211753_s_at), score: 0.82 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.8 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: 0.78 S100A14S100 calcium binding protein A14 (218677_at), score: 0.85 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.83 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.83 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.88 TBX21T-box 21 (220684_at), score: 0.79 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 1 TRA@T cell receptor alpha locus (216540_at), score: 0.89 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |