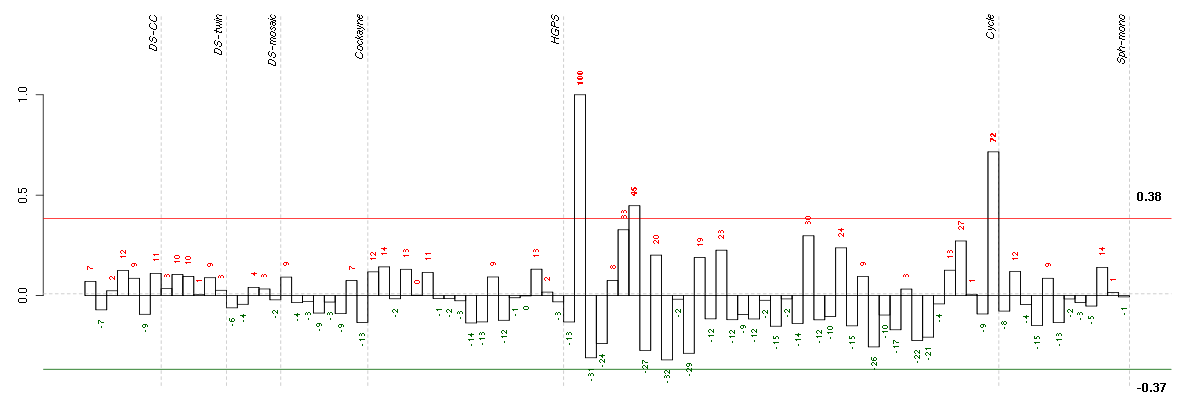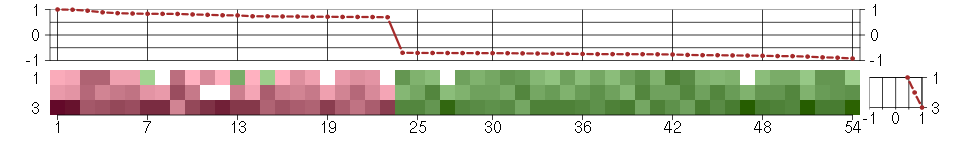

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 1 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.7 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.77 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: -0.71 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.83 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.83 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.73 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.73 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.86 CEP164centrosomal protein 164kDa (204251_s_at), score: -0.85 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.8 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.88 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.75 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: -0.71 EPS8L1EPS8-like 1 (218778_x_at), score: 0.84 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.75 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.77 FKRPfukutin related protein (219853_at), score: -0.74 FOXK2forkhead box K2 (203064_s_at), score: -0.78 FXYD2FXYD domain containing ion transport regulator 2 (207434_s_at), score: 0.72 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.72 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.79 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.76 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.81 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: 0.7 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.83 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.73 IRF2interferon regulatory factor 2 (203275_at), score: -0.79 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: 0.89 KIAA1305KIAA1305 (220911_s_at), score: -0.71 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.99 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: 0.83 LOC80054hypothetical LOC80054 (220465_at), score: 0.7 NEFMneurofilament, medium polypeptide (205113_at), score: 0.95 NEO1neogenin homolog 1 (chicken) (204321_at), score: -0.71 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.71 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.71 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.89 PHF2PHD finger protein 2 (212726_at), score: -0.92 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.75 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.72 RBM38RNA binding motif protein 38 (212430_at), score: 0.73 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.71 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.81 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.7 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.79 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.76 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.83 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.72 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: -0.74 TRIM21tripartite motif-containing 21 (204804_at), score: -0.73 TTC30Atetratricopeptide repeat domain 30A (213679_at), score: -0.81 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.75 ZNF3zinc finger protein 3 (212684_at), score: -0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |