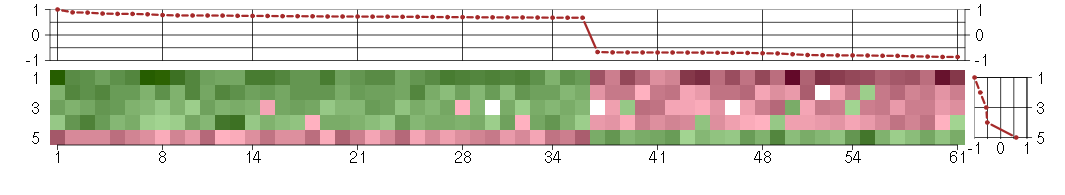

Under-expression is coded with green,
over-expression with red color.



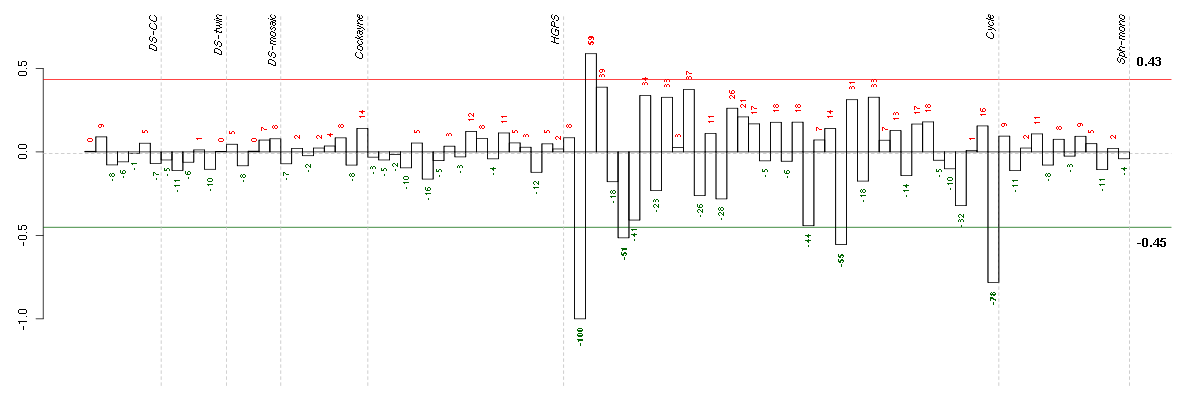
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.76 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.74 AMPD3adenosine monophosphate deaminase (isoform E) (207992_s_at), score: 0.69 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: 0.7 ATMataxia telangiectasia mutated (210858_x_at), score: -0.7 ATN1atrophin 1 (40489_at), score: 0.73 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.72 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.67 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.69 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.69 CCDC40coiled-coil domain containing 40 (220592_at), score: -0.78 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.68 CEP164centrosomal protein 164kDa (204251_s_at), score: 0.81 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.72 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.72 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.75 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.88 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: 0.71 EPS8L1EPS8-like 1 (218778_x_at), score: -0.7 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.73 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.86 FKRPfukutin related protein (219853_at), score: 0.83 FOXK2forkhead box K2 (203064_s_at), score: 0.84 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.8 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.76 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.68 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.79 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.68 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 1 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.69 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.69 IRF2interferon regulatory factor 2 (203275_at), score: 0.75 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.84 KIAA1305KIAA1305 (220911_s_at), score: 0.71 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.86 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.83 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.72 NEFMneurofilament, medium polypeptide (205113_at), score: -0.82 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.7 NUPL2nucleoporin like 2 (204003_s_at), score: -0.69 NXPH3neurexophilin 3 (221991_at), score: -0.81 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.73 PHF2PHD finger protein 2 (212726_at), score: 0.78 PXNpaxillin (211823_s_at), score: 0.76 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.69 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.8 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.69 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: 0.68 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.77 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.89 SLC22A5solute carrier family 22 (organic cation/carnitine transporter), member 5 (205074_at), score: 0.71 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.68 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.77 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.68 TBX2T-box 2 (40560_at), score: 0.74 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.8 TSKUtsukushin (218245_at), score: 0.75 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.7 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.69 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |