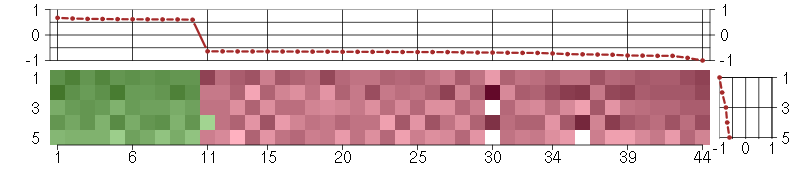

Under-expression is coded with green,
over-expression with red color.



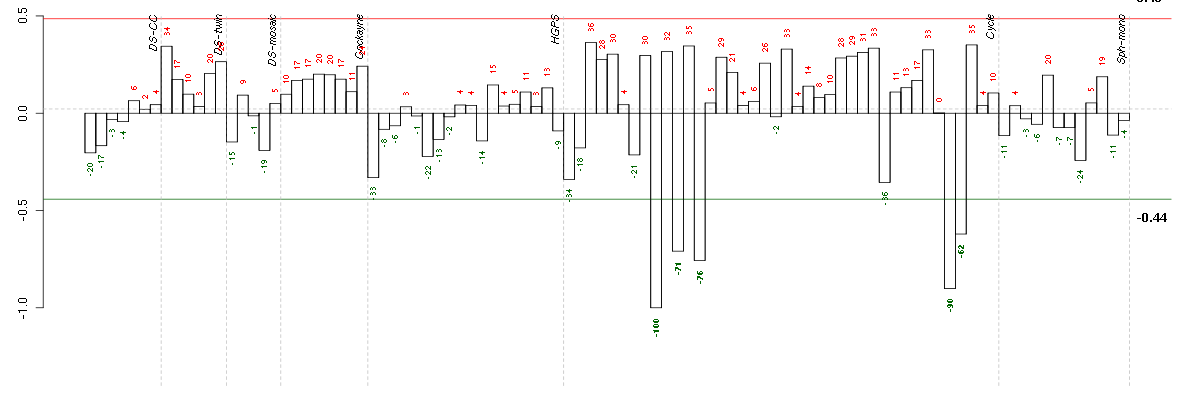
ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.65 AHI1Abelson helper integration site 1 (221569_at), score: -0.66 AREGamphiregulin (205239_at), score: -0.69 BMP6bone morphogenetic protein 6 (206176_at), score: -1 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.66 CBR3carbonyl reductase 3 (205379_at), score: 0.62 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.63 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.6 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.66 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.68 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.64 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: -0.77 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.67 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.81 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: -0.67 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.69 GKglycerol kinase (207387_s_at), score: -0.82 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.9 GPR183G protein-coupled receptor 183 (205419_at), score: -0.76 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.66 H1FXH1 histone family, member X (204805_s_at), score: 0.62 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.8 ITGB3BPintegrin beta 3 binding protein (beta3-endonexin) (205176_s_at), score: 0.61 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.65 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.75 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.67 MFAP3microfibrillar-associated protein 3 (213123_at), score: -0.7 NCALDneurocalcin delta (211685_s_at), score: -0.64 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.61 NOX4NADPH oxidase 4 (219773_at), score: -0.65 OTUB2OTU domain, ubiquitin aldehyde binding 2 (219369_s_at), score: -0.67 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (203879_at), score: -0.65 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.82 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.61 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.65 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.7 SMAD3SMAD family member 3 (218284_at), score: 0.63 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.65 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.72 TMEM2transmembrane protein 2 (218113_at), score: -0.65 TOM1target of myb1 (chicken) (202807_s_at), score: -0.65 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.7 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.77 ZNF395zinc finger protein 395 (218149_s_at), score: 0.67
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |