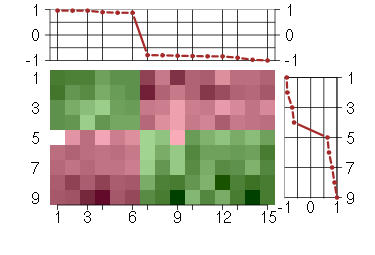

Under-expression is coded with green,
over-expression with red color.



ANGangiogenin, ribonuclease, RNase A family, 5 (213397_x_at), score: -0.84 BNIP3LBCL2/adenovirus E1B 19kDa interacting protein 3-like (221479_s_at), score: -0.8 CTSOcathepsin O (203758_at), score: -0.84 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: 0.89 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: -0.81 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.97 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: -0.82 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: 0.87 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.79 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: 0.95 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: 0.96 RHOQras homolog gene family, member Q (212119_at), score: -0.89 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.87 TOE1target of EGR1, member 1 (nuclear) (204080_at), score: 0.95 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -1
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |