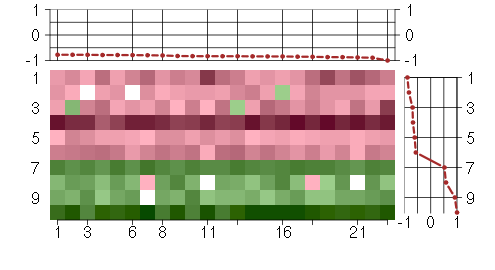

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
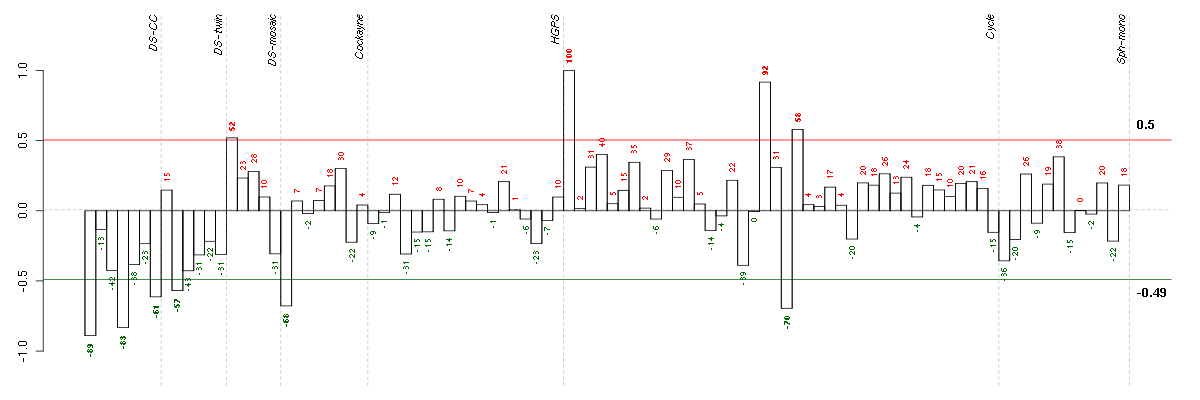
BTNL3butyrophilin-like 3 (217207_s_at), score: -0.82 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.8 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.84 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.83 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.77 DRD5dopamine receptor D5 (208486_at), score: -0.79 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.83 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.78 LHX3LIM homeobox 3 (221670_s_at), score: -1 MMRN2multimerin 2 (219091_s_at), score: -0.87 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.79 NRN1neuritin 1 (218625_at), score: -0.79 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.84 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.89 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.83 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.83 RETret proto-oncogene (205879_x_at), score: -0.79 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.85 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: -0.84 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.78 TBX21T-box 21 (220684_at), score: -0.87 TRA@T cell receptor alpha locus (216540_at), score: -0.85
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |