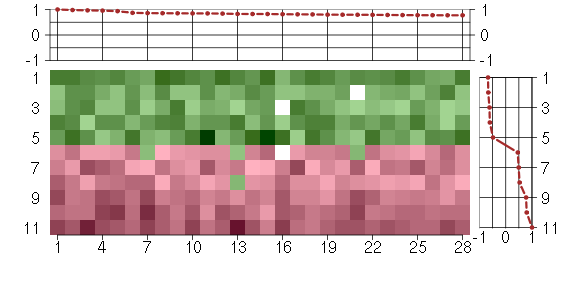

Under-expression is coded with green,
over-expression with red color.



ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.97 AHI1Abelson helper integration site 1 (221569_at), score: 0.79 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: 0.78 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.8 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.85 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.96 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.84 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.81 EVI5ecotropic viral integration site 5 (209717_at), score: 0.77 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.87 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.81 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 1 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.82 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.8 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.86 LOC391132similar to hCG2041276 (216177_at), score: 0.77 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.81 MYO1Bmyosin IB (212364_at), score: 0.85 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.78 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.79 RNF11ring finger protein 11 (208924_at), score: 0.82 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.78 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.93 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.85 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.98 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.85 YTHDC2YTH domain containing 2 (213077_at), score: 0.79 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.83
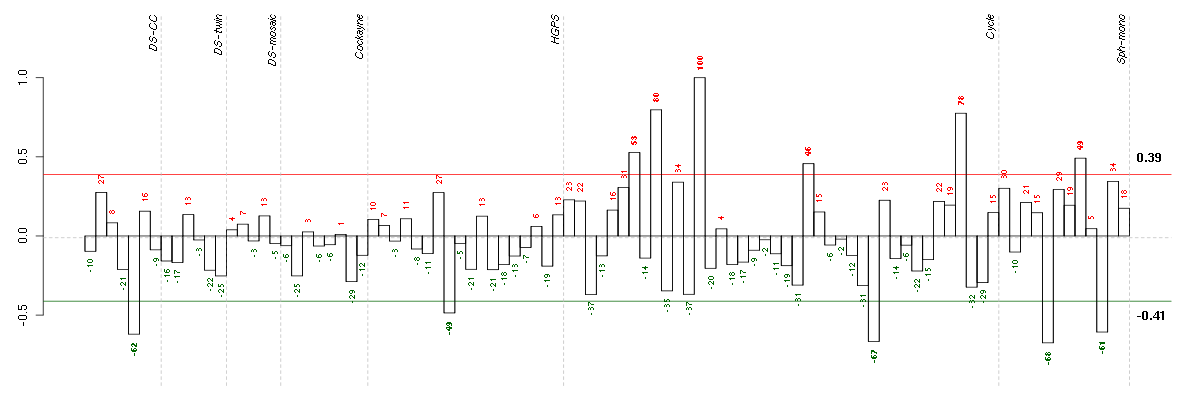
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |