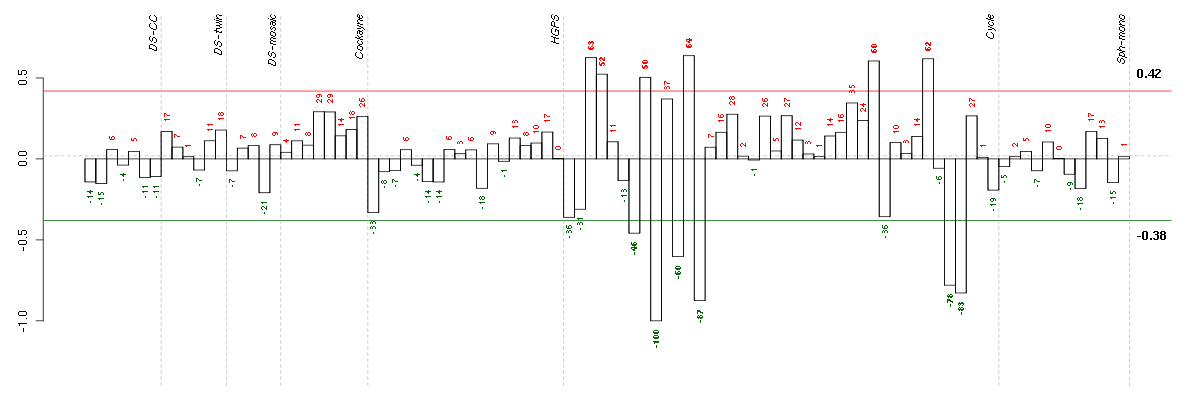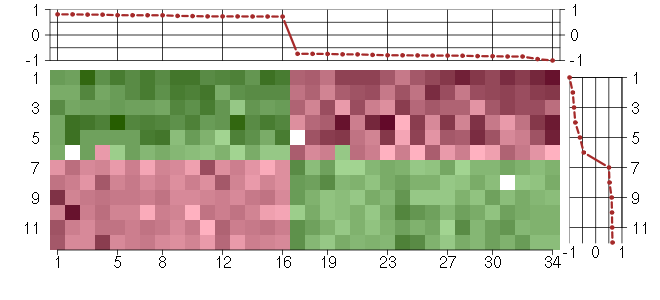

Under-expression is coded with green,
over-expression with red color.

skeletal system development
The process whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure. The skeleton is the bony framework of the body in vertebrates (endoskeleton) or the hard outer envelope of insects (exoskeleton or dermoskeleton).
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
osteoblast differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, the mesodermal cell that gives rise to bone.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
osteoblast differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, the mesodermal cell that gives rise to bone.
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.


ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.76 AIM1absent in melanoma 1 (212543_at), score: 0.75 BMP6bone morphogenetic protein 6 (206176_at), score: -1 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.74 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.72 CALB2calbindin 2 (205428_s_at), score: -0.74 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.77 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.77 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.77 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.84 ETV1ets variant 1 (221911_at), score: 0.73 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.8 GKglycerol kinase (207387_s_at), score: -0.81 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.95 H1FXH1 histone family, member X (204805_s_at), score: 0.74 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.76 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.84 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.79 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.74 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.72 NOX4NADPH oxidase 4 (219773_at), score: -0.81 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.81 PCDH9protocadherin 9 (219737_s_at), score: -0.81 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.73 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.73 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.83 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.8 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.78 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.83 SMAD3SMAD family member 3 (218284_at), score: 0.8 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.81 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.78 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.73 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.8
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |