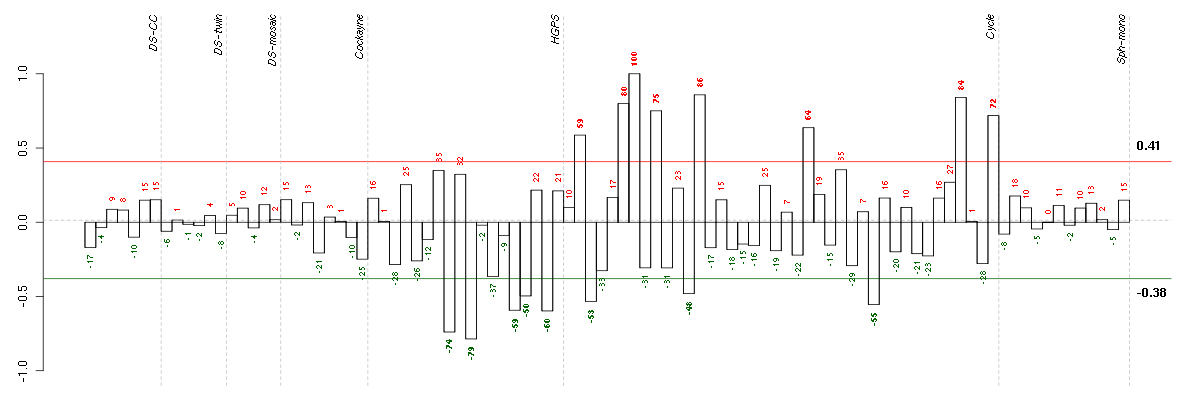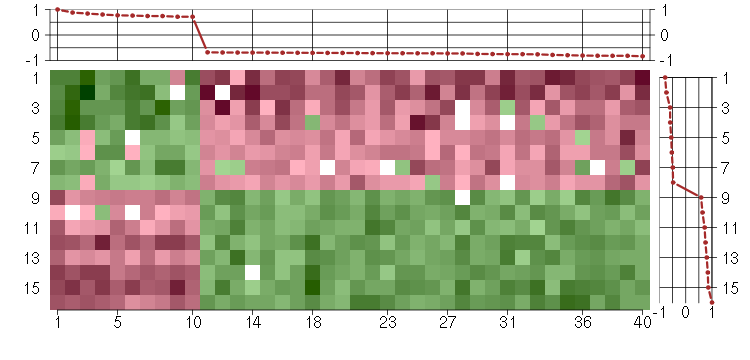

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
homophilic cell adhesion
The attachment of an adhesion molecule in one cell to an identical molecule in an adjacent cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.


BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: -0.7 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.69 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.7 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: 0.74 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.72 CICcapicua homolog (Drosophila) (212784_at), score: -0.71 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.75 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.75 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.72 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.74 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.7 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: -0.69 DNM1dynamin 1 (215116_s_at), score: -0.72 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.79 FOXK2forkhead box K2 (203064_s_at), score: -0.71 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: -0.7 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: -0.81 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.8 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.72 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.75 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.84 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.88 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.84 NEFMneurofilament, medium polypeptide (205113_at), score: 1 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.76 NRXN3neurexin 3 (205795_at), score: 0.76 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.82 PARVBparvin, beta (37965_at), score: -0.72 PCDH9protocadherin 9 (219737_s_at), score: 0.72 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.68 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.71 RNF220ring finger protein 220 (219988_s_at), score: -0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.77 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.82 SDC1syndecan 1 (201287_s_at), score: -0.7 SF1splicing factor 1 (208313_s_at), score: -0.81 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.78 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.74 TMSB15Bthymosin beta 15B (214051_at), score: -0.69 ZNF536zinc finger protein 536 (206403_at), score: -0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |