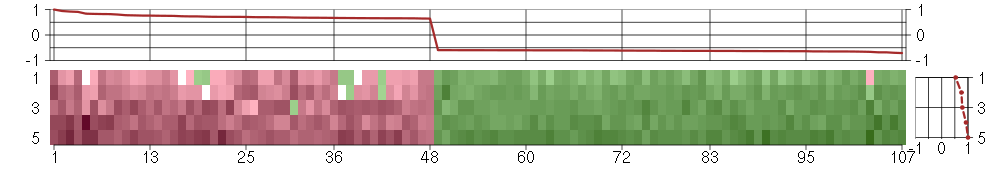

Under-expression is coded with green,
over-expression with red color.

DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
cell cycle checkpoint
The cell cycle regulatory process by which progression through the cycle can be halted until conditions are suitable for the cell to proceed to the next stage.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
DNA-dependent DNA replication
The process whereby new strands of DNA are synthesized, using parental DNA as a template for the DNA-dependent DNA polymerases that synthesize the new strands.
organelle organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
microtubule-based process
Any cellular process that depends upon or alters the microtubule cytoskeleton, that part of the cytoskeleton comprising microtubules and their associated proteins.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitotic cell cycle checkpoint
A signal transduction-based surveillance mechanism that ensures accurate chromosome replication and segregation by preventing progression through a mitotic cell cycle until conditions are suitable for the cell to proceed to the next stage.
mitotic cell cycle spindle assembly checkpoint
A signal transduction based surveillance mechanism that ensures the fidelity of cell division by preventing the premature advance of cells from metaphase to anaphase prior to the successful attachment of kinetochores to spindle microtubules (spindle assembly).
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
spindle checkpoint
A cell cycle checkpoint that delays the metaphase/anaphase transition until the spindle is correctly assembled and chromosomes are attached to the spindle.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
organelle fission
The creation of two or more organelles by division of one organelle.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
S phase
A cell cycle process comprising the steps by which a cell progresses through S phase, the part of the cell cycle during which DNA synthesis takes place.
interphase
A cell cycle process comprising the steps by which a cell progresses through interphase, the stage of cell cycle between successive rounds of chromosome segregation. Canonically, interphase is the stage of the cell cycle during which the biochemical and physiologic functions of the cell are performed and replication of chromatin occurs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitotic cell cycle checkpoint
A signal transduction-based surveillance mechanism that ensures accurate chromosome replication and segregation by preventing progression through a mitotic cell cycle until conditions are suitable for the cell to proceed to the next stage.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
S phase
A cell cycle process comprising the steps by which a cell progresses through S phase, the part of the cell cycle during which DNA synthesis takes place.
DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
mitotic cell cycle spindle assembly checkpoint
A signal transduction based surveillance mechanism that ensures the fidelity of cell division by preventing the premature advance of cells from metaphase to anaphase prior to the successful attachment of kinetochores to spindle microtubules (spindle assembly).
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.

nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
condensed chromosome
A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct structure.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosome, centromeric region
The region of a chromosome that includes the centromere and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
chromatin
The ordered and organized complex of DNA and protein that forms the chromosome.
spindle pole
Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
spindle pole
Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.

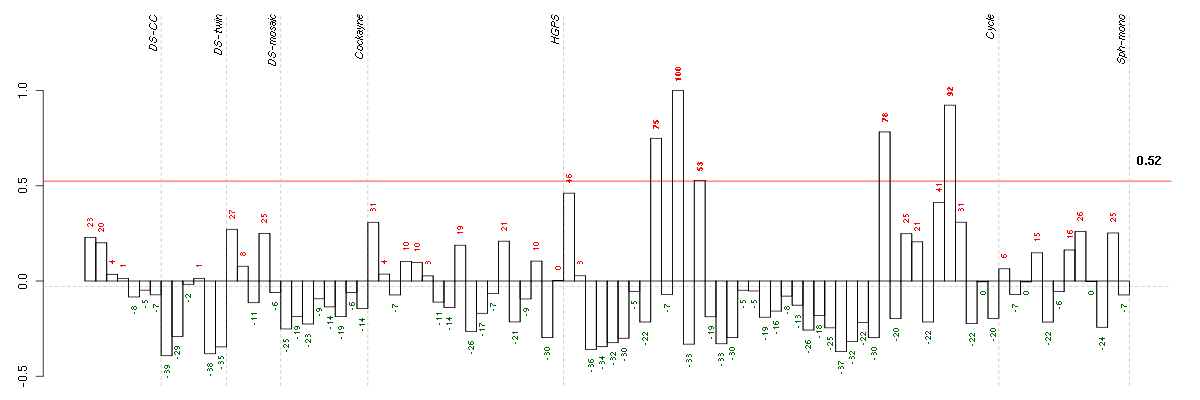
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 03030 | 4.656e-06 | 0.5033 | 8 | 33 | DNA replication |
| 04110 | 7.906e-03 | 1.495 | 8 | 98 | Cell cycle |
ANGPTL4angiopoietin-like 4 (221009_s_at), score: 0.66 AREGamphiregulin (205239_at), score: 0.84 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.65 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (205704_s_at), score: 0.66 AURKBaurora kinase B (209464_at), score: -0.64 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: 0.72 BMP2bone morphogenetic protein 2 (205289_at), score: 0.83 BMP6bone morphogenetic protein 6 (206176_at), score: 1 BRCA2breast cancer 2, early onset (208368_s_at), score: -0.62 BUB1budding uninhibited by benzimidazoles 1 homolog (yeast) (209642_at), score: -0.6 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (203755_at), score: -0.6 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: 0.65 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.76 C18orf24chromosome 18 open reading frame 24 (217640_x_at), score: -0.65 CBR3carbonyl reductase 3 (205379_at), score: -0.7 CCNB2cyclin B2 (202705_at), score: -0.6 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.62 CDCA3cell division cycle associated 3 (221436_s_at), score: -0.6 CDCA8cell division cycle associated 8 (221520_s_at), score: -0.59 CDYLchromodomain protein, Y-like (203100_s_at), score: 0.65 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: -0.65 CENPIcentromere protein I (214804_at), score: -0.64 CENPMcentromere protein M (218741_at), score: -0.62 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.6 CKAP2cytoskeleton associated protein 2 (218252_at), score: -0.62 DEPDC1DEP domain containing 1 (220295_x_at), score: -0.61 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: 0.64 E2F8E2F transcription factor 8 (219990_at), score: -0.63 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: 0.94 EZH2enhancer of zeste homolog 2 (Drosophila) (203358_s_at), score: -0.63 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.67 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: -0.61 FANCGFanconi anemia, complementation group G (203564_at), score: -0.6 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: 0.7 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: 0.76 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: 0.75 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.65 GALNT4UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) (220442_at), score: 0.71 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: 0.76 GINS2GINS complex subunit 2 (Psf2 homolog) (221521_s_at), score: -0.6 GKglycerol kinase (207387_s_at), score: 0.71 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.68 GPR183G protein-coupled receptor 183 (205419_at), score: 0.92 GPSM2G-protein signaling modulator 2 (AGS3-like, C. elegans) (221922_at), score: -0.59 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: -0.6 HELLShelicase, lymphoid-specific (220085_at), score: -0.6 HJURPHolliday junction recognition protein (218726_at), score: -0.62 HMGB2high-mobility group box 2 (208808_s_at), score: -0.63 HMMRhyaluronan-mediated motility receptor (RHAMM) (207165_at), score: -0.63 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: 0.82 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: 0.71 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.69 IL1Binterleukin 1, beta (39402_at), score: 0.67 IL8interleukin 8 (211506_s_at), score: 0.7 ITGB3BPintegrin beta 3 binding protein (beta3-endonexin) (205176_s_at), score: -0.65 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: 0.82 KIF11kinesin family member 11 (204444_at), score: -0.61 KIF15kinesin family member 15 (219306_at), score: -0.63 KIF18Bkinesin family member 18B (222039_at), score: -0.6 KIF22kinesin family member 22 (202183_s_at), score: -0.6 KIF4Akinesin family member 4A (218355_at), score: -0.63 LAMC2laminin, gamma 2 (202267_at), score: 0.73 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: 0.71 MCM2minichromosome maintenance complex component 2 (202107_s_at), score: -0.61 MCM5minichromosome maintenance complex component 5 (216237_s_at), score: -0.6 MCM7minichromosome maintenance complex component 7 (210983_s_at), score: -0.6 MFAP3microfibrillar-associated protein 3 (213123_at), score: 0.67 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: -0.6 MLF1IPMLF1 interacting protein (218883_s_at), score: -0.61 MLXIPMLX interacting protein (202519_at), score: 0.68 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.66 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: -0.63 NEIL3nei endonuclease VIII-like 3 (E. coli) (219502_at), score: -0.61 NUSAP1nucleolar and spindle associated protein 1 (218039_at), score: -0.6 OIP5Opa interacting protein 5 (213599_at), score: -0.62 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (203879_at), score: 0.78 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: 0.73 POLA1polymerase (DNA directed), alpha 1, catalytic subunit (204835_at), score: -0.68 POLD1polymerase (DNA directed), delta 1, catalytic subunit 125kDa (203422_at), score: -0.61 PRIM1primase, DNA, polypeptide 1 (49kDa) (205053_at), score: -0.63 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.91 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.77 RFC3replication factor C (activator 1) 3, 38kDa (204128_s_at), score: -0.64 RNASEH2Aribonuclease H2, subunit A (203022_at), score: -0.61 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: 0.66 SMC2structural maintenance of chromosomes 2 (204240_s_at), score: -0.62 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.66 SPATA2Lspermatogenesis associated 2-like (214965_at), score: 0.66 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (209891_at), score: -0.6 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.71 TACC3transforming, acidic coiled-coil containing protein 3 (218308_at), score: -0.62 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.7 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.59 THBDthrombomodulin (203887_s_at), score: 0.67 TIMELESStimeless homolog (Drosophila) (203046_s_at), score: -0.62 TMEM194Atransmembrane protein 194A (212621_at), score: -0.71 TOM1target of myb1 (chicken) (202807_s_at), score: 0.68 TPX2TPX2, microtubule-associated, homolog (Xenopus laevis) (210052_s_at), score: -0.61 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: 0.8 TTC17tetratricopeptide repeat domain 17 (218972_at), score: 0.73 TTKTTK protein kinase (204822_at), score: -0.63 VCLvinculin (200930_s_at), score: 0.69 WEE1WEE1 homolog (S. pombe) (212533_at), score: -0.66 YRDCyrdC domain containing (E. coli) (218647_s_at), score: 0.75 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: -0.65 ZNF222zinc finger protein 222 (206175_x_at), score: 0.68 ZNF395zinc finger protein 395 (218149_s_at), score: -0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |