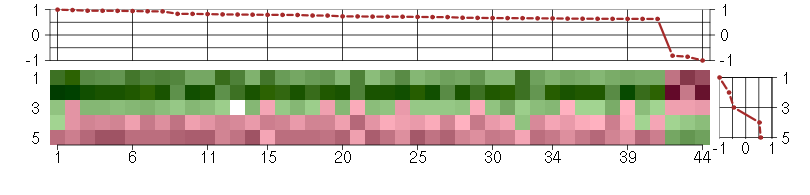

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ADAM23ADAM metallopeptidase domain 23 (206046_at), score: 0.71 AGRNagrin (212285_s_at), score: 0.63 APOL1apolipoprotein L, 1 (209546_s_at), score: 0.76 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.79 CA11carbonic anhydrase XI (209726_at), score: 0.93 CFIcomplement factor I (203854_at), score: 0.73 CLDN1claudin 1 (218182_s_at), score: 0.63 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.66 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: 0.8 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: 0.94 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.66 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: 0.98 FOXF1forkhead box F1 (205935_at), score: 0.63 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.66 GPC4glypican 4 (204983_s_at), score: 0.83 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.65 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.95 HOXB9homeobox B9 (216417_x_at), score: -0.85 HSF1heat shock transcription factor 1 (202344_at), score: 0.64 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.8 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.64 LAMA5laminin, alpha 5 (210150_s_at), score: 0.7 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.67 MSR1macrophage scavenger receptor 1 (208423_s_at), score: -0.81 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.65 NINJ1ninjurin 1 (203045_at), score: 0.78 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.63 PAPPA2pappalysin 2 (213332_at), score: 0.79 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: -1 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.82 PKP2plakophilin 2 (207717_s_at), score: 0.95 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.64 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.93 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: 0.81 SCARB1scavenger receptor class B, member 1 (201819_at), score: 0.95 SCG2secretogranin II (chromogranin C) (204035_at), score: 0.99 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.7 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.68 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: 0.83 TMEM132Atransmembrane protein 132A (218834_s_at), score: 0.72 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.73 TPD52tumor protein D52 (201690_s_at), score: 0.72 ZNF35zinc finger protein 35 (206096_at), score: 0.72 ZNF711zinc finger protein 711 (207781_s_at), score: 0.76
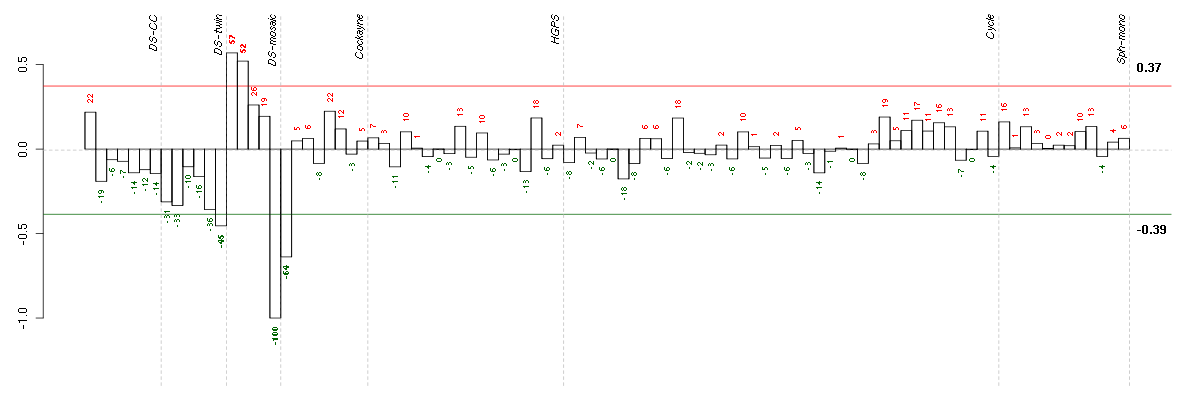
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |